Contents
What is shRNA (Short-hairpin RNA)?
The shRNA is our short hairpin RNA, which is shorter, artificially manufactured, double-stranded ribonucleic acid that can be used in gene silencing investigations.
- shRNA is a form of RNA interference (RNAi) technology used to investigate the function of uncharacterized genes.
- RNAi functions by inhibiting gene function in order to examine the affected processes. It is utilised to characterise biological pathways by identifying interactions between genes, genes, and proteins.
- It also contributes to the advancement of our knowledge of the molecular pathways underlying disease states.
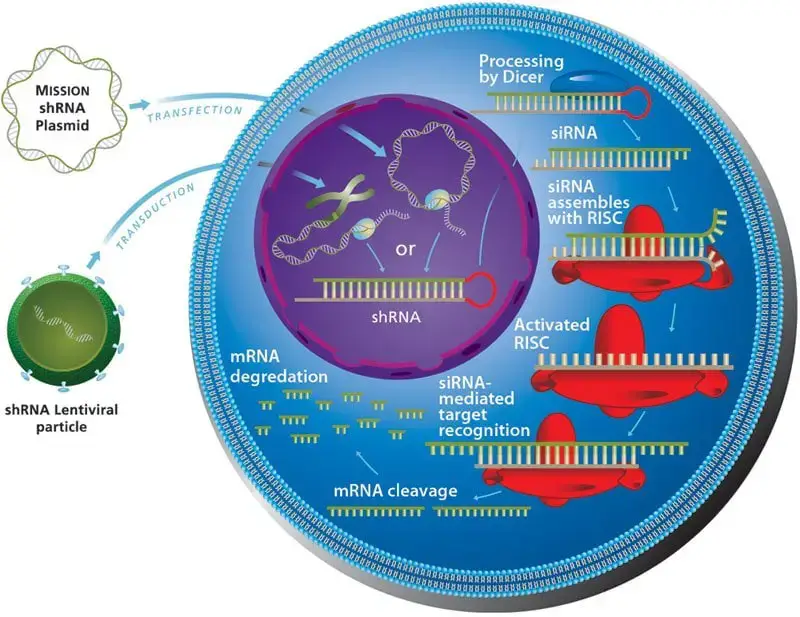
- Experimentally, RNA interference (RNAi) can be induced with small interfering RNA (siRNA) duplexes or with shRNA expressed from either a plasmid vector or genomic DNA after lentiviral-mediated integration.
- shRNAs can be administered to cells by lipid transfection of plasmid vectors or viral transduction.
- The shRNA sequence is encoded in the delivered sequence, and nuclear expression of the shRNA is driven by either an RNA Pol III promoter (such as U6 or H1) or an RNA Pol II promoter (such as CMV).
- The natural Dicer enzyme cleaves the shRNA to form the required siRNA duplexes, which then connect with RISC.
- When shRNA is supplied by lentiviral vectors, the sequence encoding the shRNA is integrated into the genome, and the knockdown effect is transmitted to daughter cells, allowing for ongoing gene silencing.
- Synthetic siRNAs between 19 and 21 base pairs with 2 nucleotide overhangs can also induce RNAi.
- siRNAs can be transported to cells by lipid transfection in addition to other techniques. Once within the cell, they directly associate with RISC.
What is RNA Interference?
This mechanism inhibits genes through the targeting of messenger RNA molecules. mRNA is degraded, blocking protein synthesis and transcription. This is accomplished by introducing double-stranded (ds) RNA into the cell, which is subsequently cleaved and bound by the RNA-induced silencing complex (RISC), which ultimately destroys the messenger RNA (mRNA) in a sequence-specific way utilising a small interfering RNA (siRNA) as a guide.
Structure of shRNA (Short-hairpin RNA)
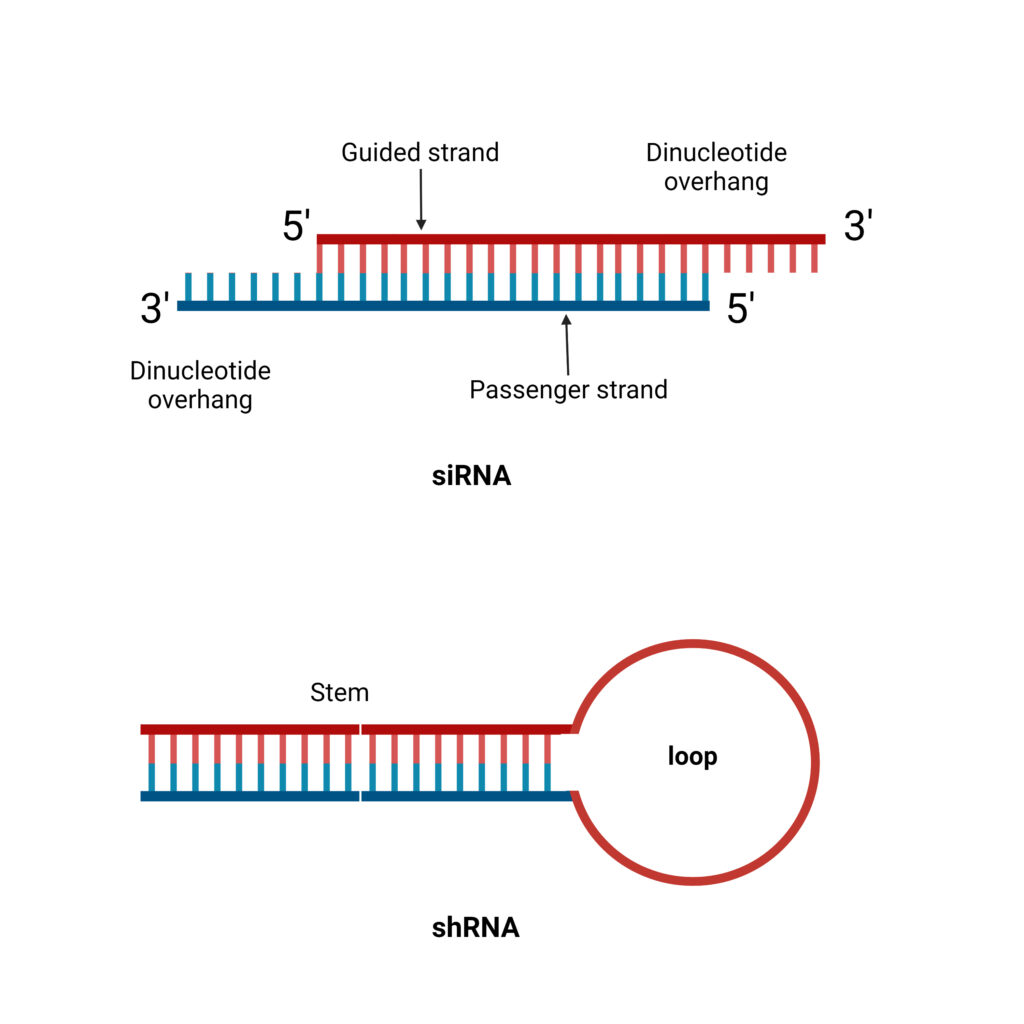
- shRNA is a 20 to 25 bp RNA polynucleotide chain in which 4 to 11 nucleotides create a hairpin-like loop that binds to the mRNA molecule.
- Unlike siRNA, it lacks the dinucleotide overhang at the 3′ OH terminus.
- During translocation, it provides a recognition site or binding motif for RNA-binding protein and prevents RNA destruction.
- A secondary RNA structural loop aids in RNA folding.
- This double-stranded molecule is utilised by scientists for several objectives, which will be discussed later in this article. But first, we must understand how the RISC and RNA interference processes operate.
- Scientists employ two forms of shRNA from a technical standpoint: plain stem-loop (hairpin) RNA and microRNA-adapted shRNA. Both have distinct compositions, structures, and functions.
- A basic stem-loop RNA consists of a 19- to 29-bp stem-loop region, a 50- to 70-nucleotide transcript, and a dinucleotide overhang at the 3′ end. It functions similarly to pre-microRNA in RNA processing. This utilises RNA polymerase III promoters.
- In contrast, the microRNA-adapted shRNA is larger than the standard one. It is at least 250 nucleotides in length. It contains natural miRNA-like mismatches, which explains why it resembles native miRNA more closely.
- Additionally, it has microRNA-like sequences on both ends.
- The shRNA is indeed a single-stranded RNA, but through intermolecular base-pairing, it produces a double-stranded structure. The guided strand contains the exact same sequence as the target mRNA, which ranges in length from 21 to 29 nucleotides. And an antisense strand that complements the guided strand correctly. This interaction forms the structure of ds-shRNA.
What is RISC (RNA interfering silencing complex)?
- The RNA interference silencing complex is composed of dicer, siRNA or shRNA, the argonaute (Ago) protein, and the dsRNA-binding protein. shRNA, like siRNA, consists of a directed strand and a passenger strand.
- Once the RISC detects the dsRNA, it recognises the directed strand and localises it on the mRNA strand that is complementary to it.
- The ‘Ago’ protein present in the complex cleaves the hybrid in the middle, followed by endo and exonuclease cleavages of the same mRNA.
- The mRNA degrades into smaller fragments and cannot be translated further. Consequently, the gene product that may be transcribed by mRNA cannot form. The short hairpin RNA can be generated artificially and employed in vivo.
- plasmid, bacterial vector, or viral vectors can be used to promote the expression of shRNA. However, viral vectors are not advised due to their infectiousness.
- Due to the low mRNA degradation rate and high turnover rate, it is evident that shRNA is more effective than siRNA.
- The shRNA can be generated in vivo using a specifically designed expression vector ( in a cell). Expression vectors are an excellent alternative for enhancing the shRNA-mediated synthetic process.
- For robust and efficient shRNA expression in a cell, the expression vector must contain the shRNA gene and the promotor sequence.
- In shRNA-mediated RNA interference, the RNA pol III or DNA pol II promoters are commonly employed. The RNA pol III is highly suggested because its shRNA expression rate is higher.
- Commonly, the U6 and H1 promoters are used in expression vectors for specific binding of RNA pol III in order to utilise RNA pol III. Unfortunately, promoters such as CMV and EF1 are not particularly efficient for shRNA expression.
- (These are all technical matters! You only need to be aware of it; don’t worry if you can’t comprehend!)
- Once the expression vector has been produced, it is introduced into the host or target genome, where the shRNA gene is transcribed by the chosen polymerase.
- Using lentivirus-mediated transduction, the entire procedure is as follows:
- shRNA is introduced into cells and reverse-transcribed into DNA.
- Following the integration of the gene and gene promoter into the host genome, ‘Drosha’ processes the pre-miRNA and pre-shRNA utilising polymerase.
- Immediately after, the exportin5 protein transfers the pre-shRNA to the cytoplasm of the cell. Here, pre-shRNA is transformed into mature shRNA. The dicer protein processes the siRNA-like dsRNA and cleaves it before loading it into the RISC.
- Leaving the passenger strand behind, the guided strand directs the complex to and attaches to the corresponding mRNA.
- Loaded in the RISC, the ‘Argo protein’ cleaves the mRNA and halts protein synthesis dictated by that mRNA. The mechanism of shRNA-mediated mRNA degradation is depicted in the diagram that follows.
shRNA mediated RNA interference
Detailed below are the general processes for shRNA-mediated RNA interference.
Step1
- Selection of the target gene; A minimum of two target sequences should be designed for a single gene in order to increase the efficacy of RNA interference (RNAi).
Step 2
- Construction of complementary shRNAs spanning from 19 to 27 base pairs to the target gene, with the 19bp shRNA producing the highest results.
Step 3
- In the next stage, either the oligonucleotide-based cloning method or the PCR-based cloning method is used to create the shRNA expression vector.
- Due to the precision and speed of PCR amplification, PCR-based cloning techniques are commonly employed. Learn more about PCR: A Comprehensive Introduction to the Polymerase Chain Reaction
- Here, a single mismatched base can inhibit hairpin formation, hence it is essential to choose the sequence with the highest capacity for hairpin formation.
Step 4
- In this step, promoter sequences for RNA pol III or DNA pol II are chosen for shRNA polymerization.
- Note: The lentivirus-mediated transduction is less dangerous than the AAS virus-mediated therapy, and it can infect both diving and non-diving cells well, which is another reason to use it.
Step 5
- After all of that, the vectors with the shRNA for the gene knockdown study are put into the cells or the target cells. Quantitative PCR analysis is used to figure out how well an infection spreads.
- Quantitative PCR, also called reverse transcription PCR, is a way to measure how much a gene is being used. Also, if the biological function of the target gene is known, its effect can be studied, such as whether the phenotype is suppressed or overexpressed.
- The use of retroviruses is not a good idea, so bacteria are now being used as vectors.
Mechanism of shRNA (Short-hairpin RNA)
- Once the vector has been added to the host genome, polymerase II or polymerase III, depending on the choice of promoter, copies the shRNA in the nucleus.
- The product is made by Drosha and acts like pri-microRNA (pri-miRNA).
- Exportin 5 takes the pre-shRNA out of the cell’s nucleus.
- Then, Dicer works on this product and adds it to the RNA-induced silencing complex (RISC).
- The sense strand (passenger) is broken.
- The antisense (guide) strand tells RISC to go to mRNA that has a sequence that matches the guide strand.
- When the two parts are a perfect match, RISC cuts the mRNA. When there isn’t a perfect match, RISC stops the mRNA from being translated.
- In both of these situations, the shRNA turns off the target gene.
shRNA design
shRNAs are usually made in one of two ways: with a simple stem-loop or with a microRNA-adapted format.
Simple stem-loop shRNA
- Basic shRNAs are based on pre-miRNA and are cloned into viral vectors, where they are copied by RNA Polymerase III (Pol III) promoters.
- shRNAs are made as single-strand molecules that are 50–70 nucleotides long and have stem loop structures. The stem is a 19–29 base pair section of double-stranded RNA that is connected to the loop by a section of single-stranded RNA and a short 3′ overhang.
- Once transcribed, shRNAs leave the nucleus, where they are cut at the loop by the nuclease Dicer in the cytoplasm. They then go into the RISC, where they tell the nuclease to cut complementary mRNA and get rid of it.
microRNA adapted shRNA
- A microRNA-adapted shRNA is made up of a shRNA stem structure with microRNA-like mismatches and the loop and flanking sequence of an endogenous microRNA.
- microRNA-adapted shRNAs are copied from RNA Polymerase II (Pol II) promoters, cut by the endogenous RNase III Drosha enzyme in the nucleus, and then sent to the cytoplasm where they are processed by Dicer and loaded into the RISC complex.
- Studies have shown that using a microRNA scaffold that can be broken down by both Drosha and Dicer could make processing more efficient and less harmful for in vivo RNAi.
shRNA applications
- shRNA makes it possible to silence genes for a long time.
- Transduction of viral-based shRNA gives access to cells that are hard to transfect with traditional cationic lipid-based strategies, such as primary and neuronal cells.
- Using pools of silencing constructs, shRNAs made from viruses have also been used to look at how genes work across the whole genome.
- Pooled or arrayed screens are now widely used to do high-throughput screens to find genes needed for things like cancer cell survival and proliferation, tumour suppressor pathway components, modulators of the mammalian circadian clock, suppressors of epithelial-mesenchymal transition, host mediators of HIV-1 replication, and regulators of cell migration. Pooled RNAi screening has also been used to look for how genes work in animal models to learn more about biology in vivo.
Therapeutic applications of shRNA
- shRNAs have been used to treat prostate cancer, melanoma, and diseases that damage nerve cells. They are also being carefully studied to see if they can be used to control the activity of a target gene in other long-term diseases.
- Huang et al. used a mouse model to test how the prolyl hydroxylase-2 (PHD2) protein could be used with shRNA to treat myocardial ischemia. Molecular imaging that did not hurt the mouse showed that blocking PHD2 with shRNA made angiogenesis much better.
- The study also found that the sense and antisense fragments of shRNA are expressed in the same way when they are driven by two H1 promoters. The down-regulation of the PHD2 gene in the mouse is caused by the knocking-down effects of these sense and antisense fragments. In turn, downregulating PHD2 turns on angiogenic proteins and genes that are part of the hypoxia response pathway.
- Studies have also shown that retrovirally delivered shRNA can stop the spread of the human immunodeficiency virus type 1 (HIV-1) for longer than other HIV treatments.
- ShRNA gene therapy has also been tested on non-obese mice with weak immune systems that were injected with CD44+ CD24- cells to see if it could stop human breast tumours from growing. It is known that CD44+ CD24- cells are what cause breast tumours, so turning down CD44 can stop the growth of tumours.
- In a study done by Pham et al., it was found that a combination of CD44 RNA lentiviral vector and doxorubicin stopped the growth of tumours very well and was safe. By going after the cancer stem cells, this could be a good way to treat breast cancer.
Challenges in shRNA mediated therapies
- The cells of the patient may make a strong immune response to the shRNA: That makes a lot of sense, right? At first, the cell may see our shRNA as the foreign, harmful, and pathogenic dsDNA and trigger a strong immune response to get rid of it.
- It can be said out of control: Expression vectors have a higher expressivity, which means that more shRNA can be made. cells can’t handle it well enough.
- The wrong mRNA is being processed: The whole process should be clear and focused on the goals. The shRNA only works on the mRNA that we want it to. When it is overexpressed, a cell with a high level of RISC saturation processes the wrong mRNAs, not the target mRNA. Target-specificity can be lost if you say too much, which is another big problem.
- Effects of shRNA not on target: Off-target means that our shRNA is acting on mRNA that isn’t the one we want to change. As we’ve already said, the technology is new and not fully understood or documented yet. Because of this, there have been some reports of off-target effects.
- The therapy hasn’t been tried out enough: The therapy hasn’t been fully tested, and it might not work as planned. This can change the way other genes are expressed, too.
- Low transfectivity: This means that the shRNA therapy doesn’t work very well at transferring genes. In particular, cells that don’t divide and other immune cells can’t be transfected accurately and well. It takes months to get shRNA-positive cells ready to be used in a transplant.
- Can’t work for all types of cells: Another problem with this therapy is that it doesn’t work for all types of cells. This means that the shRNA doesn’t go after all types of cells. It can work great with some cell lines but not at all with others.
References
- Tsujiuchi, T., Miller, A. D., Wakabayashi, T., & Natsume, A. (2014). RNA Interference Therapeutics for Tumor Therapy. Gene Therapy of Cancer, 393–408. doi:10.1016/b978-0-12-394295-1.00027-5
- Moore CB, Guthrie EH, Huang MT, Taxman DJ. Short hairpin RNA (shRNA): design, delivery, and assessment of gene knockdown. Methods Mol Biol. 2010;629:141-58. doi: 10.1007/978-1-60761-657-3_10. PMID: 20387148; PMCID: PMC3679364.
- Sheng, P., Flood, K. A., & Xie, M. (2020). Short Hairpin RNAs for Strand-Specific Small Interfering RNA Production. Frontiers in Bioengineering and Biotechnology, 8. doi:10.3389/fbioe.2020.00940
- https://en.wikipedia.org/wiki/Short_hairpin_RNA
- https://geneticeducation.co.in/what-is-shrna-short-hairpin-rna/
- https://horizondiscovery.com/en/applications/rnai/shrna-applications
- https://www.news-medical.net/life-sciences/Short-Hairpin-RNA-(shRNA)-Interference-Therapeutic-Applications.aspx
- https://www.news-medical.net/life-sciences/Short-Hairpin-RNA-(shRNA)-Interference.aspx
- https://genesdev.cshlp.org/content/16/8/948
- https://mcmanuslab.ucsf.edu/node/274
- https://www.sigmaaldrich.com/IN/en/technical-documents/technical-article/genomics/gene-expression-and-silencing/shrna-process-and-shrna-diagram
- https://www.labome.com/method/siRNAs-and-shRNAs-Tools-for-Protein-Knockdown-by-Gene-Silencing.html