What are Plasmids?
- Plasmids are small, extrachromosomal DNA structures found primarily within bacterial cells, distinct from the primary chromosomal DNA. Unlike the chromosomal DNA that encompasses the essential genetic blueprint of the organism, plasmids are typically circular and replicate autonomously. Their presence is not vital for the host’s basic survival; however, they often harbor genes that confer specific advantages, such as antibiotic resistance or toxigenicity.
- These genetic elements, while predominantly observed in bacteria, have also been identified in archaea and certain eukaryotic organisms. Each bacterial cell may contain approximately 1 to 20 copies of a plasmid. Some plasmids, termed episomes, possess the capability to integrate into the bacterial chromosome and replicate alongside it.
- One of the remarkable features of plasmids is their role in horizontal gene transfer, particularly through a process known as conjugation. This mechanism facilitates the transfer of genetic material between bacteria, even those of different species, thereby promoting genetic diversity. It’s noteworthy that plasmids, despite their genetic content, are not deemed independent life forms. They lack the protective protein coat, or capsid, characteristic of viruses and do not encapsulate their genetic material for transfer. However, certain plasmids do encode for the conjugative “sex” pilus, essential for their transfer between hosts.
- In the realm of molecular biology, artificial plasmids have gained prominence as vectors for molecular cloning. These engineered plasmids drive the replication of recombinant DNA within host organisms, proving invaluable for scientific research. Furthermore, with advancements in biotechnology, synthetic plasmids can now be procured, furthering their utility in laboratory settings.
- In summary, plasmids are versatile genetic elements, pivotal in bacterial adaptation and survival, and invaluable tools in biotechnological research. Their ability to replicate independently and confer specific traits to their host underscores their significance in the microbial world.

Definition of Plasmids
Plasmids are small, circular, extrachromosomal DNA molecules found primarily in bacteria that can replicate independently of the main chromosomal DNA and often carry genes beneficial to the host organism, such as those conferring antibiotic resistance.
Properties/Characteristics of Plasmids
Bacterial plasmids are extrachromosomal DNA fragments that exist within bacterial cells. These are some of their key characteristics:
- Extrachromosomal Nature: Plasmids are distinct from the primary chromosomal DNA of bacteria. They exist outside the main chromosome and have their own unique genetic content.
- Structure: Typically, plasmids are double-stranded DNA molecules. However, exceptions exist, such as the linear plasmids observed in bacteria like Streptomyces spp and Borrelia spp.
- Independent Replication: One of the defining features of plasmids is their ability to replicate autonomously, independent of the chromosomal DNA replication.
- Beneficial Attributes: While the absence of a plasmid does not hinder the basic functioning of a bacterial cell, its presence often confers advantageous traits to the host.
- Nomenclature: Plasmids have been referred to by various names, including sex factors, conjugants, extrachromosomal replicons, and transfer factors.
- Copy Number: This term denotes the number of plasmid copies within a bacterial cell. It’s observed that smaller plasmids tend to be more abundant, whereas larger ones are fewer in number.
- Compatibility: Different plasmids have varying abilities to coexist within the same bacterial cell. This characteristic is termed as the compatibility of plasmids.
- Replication Origin: For independent replication, plasmids necessitate an origin of replication, termed a replicon. While smaller plasmids leverage the host’s replicative enzymes, larger ones might possess genes tailored for their replication.
- Episomal Nature: Certain plasmids, known as episomes in prokaryotes, have the capability to integrate into the host chromosome.
- Beneficial Genes: Plasmids frequently harbor genes that bestow beneficial traits upon the host cell. Examples include genes conferring antibiotic resistance or specific metabolic capabilities.
- Size Variation: Plasmids exhibit a wide size range. They can be as small as mini-plasmids with less than 1 kbp or as large as megaplasmids spanning several Mbp.
- Circularity: While the majority of plasmids are circular in structure, linear variants do exist and necessitate unique replication strategies.
- Copy Number Variation: The quantity of plasmids within a cell can vary extensively, from a single copy to several hundreds.
- Partition Systems: Plasmids with lower copy numbers employ partition systems to ensure even distribution to daughter cells during bacterial cell division.
In essence, bacterial plasmids are versatile genetic elements that play a pivotal role in bacterial adaptability, survival, and evolution. Their unique properties make them invaluable tools in genetic engineering and biotechnological research.
Shapes of plasmids
Plasmids, the extrachromosomal DNA fragments in bacterial cells, can exist in various conformations based on their structural attributes. The shapes of plasmids are integral to their functionality and interactions within the cell. Here are the five distinct shapes of plasmids:
- Nicked Open-Circular: In this conformation, one of the DNA strands of the plasmid has experienced a break or “nick.” This results in a structure where the DNA remains circular, but with a discontinuity in one of its strands.
- Relaxed Circular: This shape is characterized by a fully intact DNA structure without any nicks or breaks. The DNA remains in a relaxed state, forming a complete loop.
- Linear: As the name suggests, in this form, the plasmid DNA does not form a loop. Instead, it extends linearly with free ends, differing from the typical circular nature of most plasmids.
- Supercoiled: This conformation is observed when the DNA of the plasmid is not only intact but also undergoes additional twisting. This added twist results in a more compact structure, making the plasmid appear smaller than its relaxed counterpart.
- Supercoiled Denatured: This shape is a variant of the supercoiled form. It is slightly more compact, making it marginally smaller in size compared to the standard supercoiled conformation.
In summary, the diverse shapes of plasmids play a pivotal role in their biological functions, interactions, and overall dynamics within bacterial cells. Understanding these conformations is crucial for insights into plasmid biology and their applications in genetic research.
Size of Plasmid
Plasmid size is a defining characteristic that influences its functionality and applications, especially in the realm of genetic research and biotechnology. Here’s an exploration of the size of plasmids:
- Size Range: Plasmids exhibit a broad size spectrum, ranging from several kilobase pairs (kb) to multiple megabase pairs (Mb). The specific size is contingent upon the type and nature of the plasmid.
- Quantity and Size Correlation: Typically, bacterial cells contain fewer small plasmids, while a singular, larger plasmid is often present. This inverse relationship between size and quantity is a notable feature of plasmid biology.
- Functionality and Size: One of the salient attributes of plasmids is their ability to facilitate horizontal gene transfer via conjugation. This property is not strictly size-dependent but is a crucial aspect of plasmid-mediated genetic exchange.
- Replication and Inheritance: Plasmids are autonomous genetic elements capable of self-replication. During bacterial cell division, these plasmids are inherited, ensuring their presence in subsequent generations.
- Significance in Genetic Research: Plasmids of sizes typically ranging from 1kb to 2kb are frequently employed in genetic research. Their relatively compact size renders them amenable to manipulation, making them invaluable tools in genetic engineering endeavors.
- Stability: A hallmark of plasmid DNA is its remarkable stability. Whether in a purified form or within a bacterial cell, plasmids can remain stable for extended durations, preserving their genetic content.
- Versatility: Owing to their stable nature and ability to replicate independently, plasmids find applications across diverse species. They are pivotal in gene transfer experiments and have potential in gene therapy protocols.
In essence, the size of a plasmid is not just a measure of its length but is intricately linked to its functionality, stability, and applications in genetic research and biotechnological innovations.
Structure of plasmids
Plasmids are extrachromosomal DNA elements that play a pivotal role in bacterial genetics and biotechnology. Their structure is intricate, encompassing several essential components that confer specific functionalities:
- Origin of Replication (OR): The origin of replication is a distinct region on the plasmid where the process of DNA replication commences. Characterized by an adenine-thymine (A=T) rich region, this area facilitates easier strand separation during replication.
- Selectable Marker Site: This segment of the plasmid houses antibiotic resistance genes. These genes are instrumental in discerning and selecting bacterial cells that harbor the plasmid, thereby aiding in the identification of successfully transformed cells.
- Promoter Region: A crucial segment for gene expression, the promoter region is where the cellular transcriptional machinery assembles to initiate the transcription of genes present on the plasmid.
- Primer Binding Site: This is a concise single-stranded DNA sequence that plays a pivotal role in processes like DNA amplification and sequencing.
- Multiple Cloning Sites: These are specialized regions on the plasmid that encompass various sequences recognized by restriction enzymes. These enzymes can bind to and cleave these sites, facilitating the insertion of foreign DNA.
- Size Variation: Plasmids exhibit a diverse size range, spanning from 2 kilobase pairs (kb) to 200 kb. This size variability allows them to harbor different numbers of genes and regulatory elements.
- Extrachromosomal Nature: While plasmids are present within the cell, they are not essential for the basic growth and development of the organism. They operate outside the chromosomal framework and are not a mandatory component of the cellular genome.
- Transfer Genes: Many plasmids possess the TRA gene, a crucial element that facilitates the transfer of the plasmid between cells, enabling horizontal gene transfer.
- Replication and Distribution: Plasmids are equipped with their distinct origin of replication, allowing them to replicate autonomously. As the host cell divides, plasmids ensure their distribution to daughter cells, maintaining their presence across generations.
- Beneficial Genes: Beyond the replication origin, plasmids often encompass genes that confer advantages to the host, such as antibiotic resistance or toxin production. These genes can enhance the survival prospects of the host in challenging environments.
In summation, plasmids are versatile genetic elements with a structured architecture that equips them with unique functionalities. Their extrachromosomal nature, coupled with their ability to confer beneficial traits, makes them invaluable tools in genetic research and biotechnological applications.
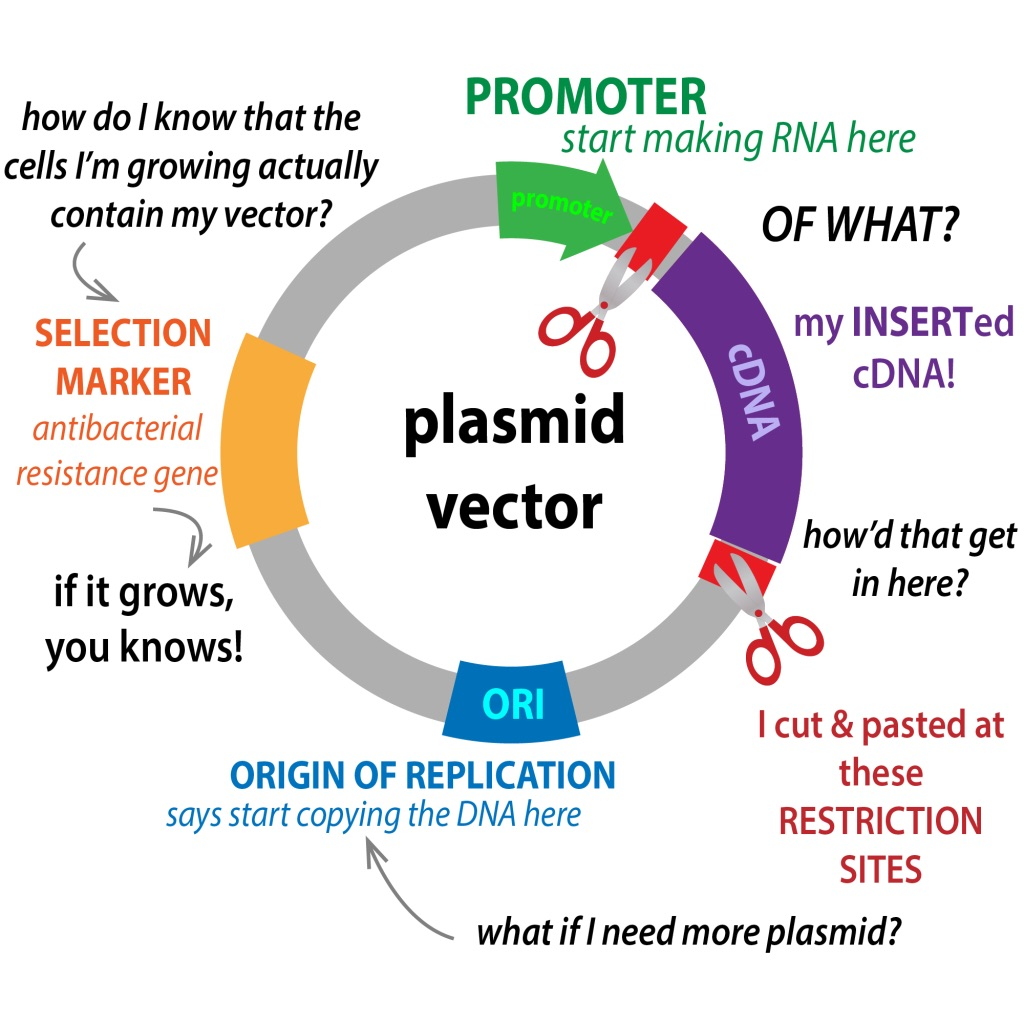
In general, plasmids are made up of three main components which comprise:
1. Origin of replication (replicon)
- The place of origin for replication (ori) is an exact location within the strand from which replication starts.
- For plasmids this area is mostly composed of A-T base pair pairs that are simpler to divide when undergoing replication.
- In comparison to the organisms’ DNA, which has many types in replication.
- Moreover, the plasmids are one of the few sources of replication since their size is smaller.
- The origin of the process, they contain several regulators that are involved in replication (e.g. Rep proteins).
2. Polylinker (multiple cloning sites)
- In the plasmid the polylinker (MCS) is among the most crucial components of the molecular. It is due to its ability to allow students to gain knowledge about the process of cloning.
- A polylinker is a brief sequence of DNA composed of several locations for cleavage through restriction enzymes. In this way, MCS allows for the simple insertion of DNA by the ligation process or digestion with restriction enzymes.
- On the site of cleavage different polylinkers are able to slice the DNA strand.
- So any of these restriction enzymes may cut the plasmid in specific places on the sire, allowing for DNA to be inserted.
3. Antibiotic resistance gene
- The gene for resistance to antibiotics is one of the primary elements of plasmids. These genes play a significant part in the development of resistance to drugs (to any or all of the antibiotics) and make treatment for certain diseases more difficult.
- Plasmids are renowned today for their capacity to transfer genes from one species or bacterium to another in the process of conjugation (contact between cells followed by the transfer of the DNA contents).
- Through this process, they can confer antimicrobial resistance to different species of bacteria. Although plasmid replication offers an advantage for bacteria (resistance to specific antibiotics) however, it can also impact the division of bacteria’s cells due to the increased burden of replication.
- This is why bacteria that have plasmids tends to be fewer populated than the ones without plasmids because of the lower rate of cell division.
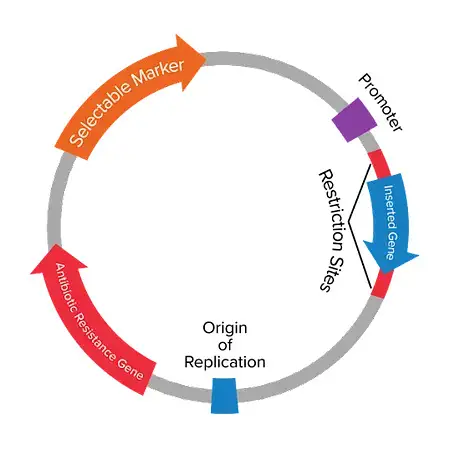
4. Some of the other components of plasmids include
- A promoter region: A promoter region is the part of plasmids responsible for recruiting the transcriptional machinery.
- Primer binding site: A primer binding site is a small sequence of DNA that is on one strand, which is usually used to aid in the process of PCR Amplification as well as DNA sequencing.
- Selection Marker: The antibiotic resistance gene (ARG) plays a significant function in the development of resistance to drugs, that allows the selection of bacteria that have plasmids. Apart from ARGs some plasmids contain other selection markers, either in nature , or via artificial introduction.
| Vector Element | Description |
| Origin of Replication (ORI) | DNA sequence which allows initiation of replication within a plasmid by recruiting replication machinery proteins |
| Antibiotic Resistance Gene | Allows for selection of plasmid-containing bacteria. |
| Multiple Cloning Site (MCS) | Short segment of DNA which contains several restriction sites allowing for the easy insertion of DNA. In expression plasmids, the MCS is often downstream from a promoter. |
| Insert | Gene, promoter or other DNA fragment cloned into the MCS for further study. |
| Promoter Region | Drives transcription of the target gene. Vital component for expression vectors: determines which cell types the gene is expressed in and amount of recombinant protein obtained. |
| Selectable Marker | The antibiotic resistance gene allows for selection in bacteria. However, many plasmids also have selectable markers for use in other cell types. |
| Primer Binding Site | A short single-stranded DNA sequence used as an initiation point for PCR amplification or sequencing. Primers can be exploited for sequence verification of plasmids. |
The structure of plasmids can be summarized as follows:
- Essential elements:
- Origin of replication (OR): A specific region where replication begins, usually an A=T rich region that facilitates strand separation during replication.
- Selectable marker site: Contains antibiotic resistance genes that aid in identifying and selecting bacteria with the plasmid.
- Promoter region: The site where the transcriptional machinery is loaded for gene expression.
- Primer binding site: A short sequence of single-stranded DNA that enables DNA amplification and sequencing.
- Multiple cloning sites: Sequences where restriction enzymes can bind and cleave the double-stranded structure, facilitating the insertion of foreign DNA.
- Plasmid size: Plasmids can range in size from 2 kilobase pairs (kb) to 200 kb.
- Extrachromosomal element: Plasmids are not necessary for the growth and development of the cell. They exist separately from the chromosomal DNA.
- TRA gene: Most plasmids contain the TRA gene, which is responsible for transferring the plasmid from one cell to another. This gene plays a crucial role in horizontal gene transfer.
Types of plasmid based on Function
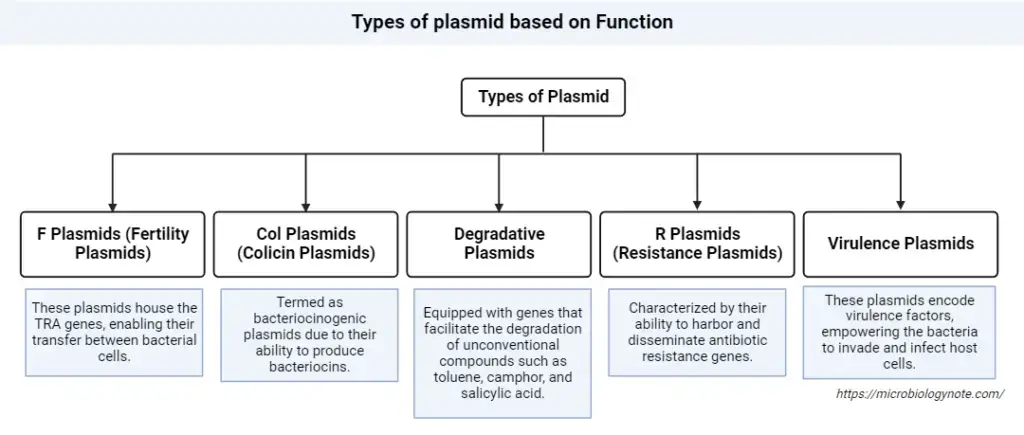
Plasmids, as extrachromosomal DNA elements, play diverse roles in bacterial physiology and adaptation. Based on their functional attributes, plasmids can be systematically classified into the following categories:
- F Plasmids (Fertility Plasmids):
- These plasmids house the TRA genes, enabling their transfer between bacterial cells.
- Capable of autonomous replication within the bacterial cell.
- They orchestrate the synthesis of a pilus, a proteinaceous structure facilitating cell-to-cell interactions.
- Additionally, they possess sequences that determine incompatibility, preventing coexistence with similar plasmids.
- R Plasmids (Resistance Plasmids):
- Characterized by their ability to harbor and disseminate antibiotic resistance genes.
- These genes confer protection against both medically relevant antibiotics and naturally occurring antibiotics in the environment.
- Typically, R plasmids are larger and exist in fewer copies within the bacterial cell.
- Col Plasmids (Colicin Plasmids):
- Termed as bacteriocinogenic plasmids due to their ability to produce bacteriocins.
- These bacteriocins can target and eliminate closely related bacterial strains that lack the Col plasmid.
- E. coli is a notable bacterium where Col plasmids have been identified.
- Degradative Plasmids:
- Equipped with genes that facilitate the degradation of unconventional compounds such as toluene, camphor, and salicylic acid.
- Bacteria harboring these plasmids can metabolize a diverse range of chemicals, aiding in environmental detoxification and resource utilization.
- Virulence Plasmids:
- These plasmids encode virulence factors, empowering the bacteria to invade and infect host cells.
- Bacterial strains with virulence plasmids can target plant, animal, and human cells, leading to various diseases.
- A quintessential example is the Ti plasmid found in Agrobacterium tumefaciens, which induces crown gall disease in plants.
In essence, the functional classification of plasmids underscores their multifaceted roles in bacterial survival, adaptation, and pathogenicity. Each category of plasmid equips the bacterium with specialized capabilities, from antibiotic resistance to metabolic versatility, shaping the ecological and pathogenic landscape of bacterial communities.
1. Fertility Plasmids (F PLASMID)
Fertility plasmids, commonly referred to as F-plasmids, are specialized genetic elements within bacteria that facilitate the process of conjugation, a mechanism of horizontal gene transfer. These plasmids are integral members of the conjugative plasmid family. Here are the salient features of F-plasmids:
- Nature and Existence:
- F-plasmids are episomes, meaning they have the capability to integrate into the bacterial chromosome or exist independently as extrachromosomal elements.
- They harbor essential genetic information that ensures their extrachromosomal survival and self-transfer capabilities.
- Role in Conjugation:
- Bacteria possessing the F-plasmid are designated as F positive (F+), acting as donors during the conjugation process. In contrast, bacteria devoid of this plasmid are termed F negative (F–) and function as recipients.
- Upon successful conjugation between an F+ and an F– bacterium, the resultant pair both emerge as F+ entities, signifying the transfer of the F-plasmid.
- Genetic Composition:
- The F-plasmid encompasses a suite of genes, including those responsible for pilin synthesis. Pilin is the foundational protein for the formation of the sex pilus, a structure instrumental in bacterial mating.
- A cluster of approximately 25 transfer (tra) genes is present in the F-plasmid. These genes orchestrate various functions, from pilus expression and DNA synthesis and transfer during mating to modulating the recipient capabilities of F+ bacteria.
- Pilus Formation:
- The pilus, synthesized under the guidance of the F-plasmid, is a proteinaceous appendage that plays a pivotal role in mediating cell-to-cell interactions, facilitating the transfer of genetic material.
- Incompatibility Sequence:
- F-plasmids also encompass specific sequences that determine incompatibility, ensuring that a singular F-plasmid type predominates within a bacterial cell.
- Significance:
- Beyond their role in conjugation, F-plasmids are emblematic of the intricate mechanisms bacteria have evolved to exchange genetic information, adapt to changing environments, and potentially acquire beneficial traits, such as antibiotic resistance.
In summary, F-plasmids are quintessential molecular tools that bacteria employ to engage in genetic exchanges, underscoring the dynamic nature of bacterial genetics and the profound implications of horizontal gene transfer in microbial evolution and adaptation.
2. Resistance Plasmids (R PLASMID)
Resistance plasmids, commonly designated as R plasmids, are specialized genetic elements within bacteria that harbor genes conferring resistance against external adversities, notably antibiotics. These plasmids play a pivotal role in the bacterial defense mechanism, enabling them to thrive in hostile environments. Here are the salient features of R plasmids:
- Nature and Composition:
- R plasmids are circular, double-stranded DNA structures.
- They can be categorized based on size into large conjugative R plasmids, which possess additional DNA for the conjugation process, and smaller non-conjugative plasmids that solely contain resistance genes.
- Components:
- R plasmids comprise two primary components:
- Resistance Transfer Factor (RTF): This segment encompasses genes responsible for intercellular transfer.
- Resistance Determinant (R-determinant): This section houses genes conferring resistance to various drugs. The transfer of drug resistance is inhibited if the RTF disassociates from the R-determinant.
- R plasmids comprise two primary components:
- Antibiotic Resistance:
- R plasmids are instrumental in transmitting antibiotic resistance genes from one bacterial cell to another, offering protection against both medically prescribed antibiotics and those naturally occurring in the environment.
- The emergence of antibiotic-resistant strains, such as certain strains of gonorrhea, underscores the significance of R plasmids in bacterial adaptation. The rapid adaptation has even necessitated shifts in antibiotic recommendations by global health organizations.
- Mechanisms of Resistance:
- R plasmids can facilitate the degradation of antibiotics or modify the bacterial membrane transport system to prevent antibiotic entry.
- They might harbor resistance genes for one or multiple drugs and can even confer resistance against specific metal ions.
- Additionally, certain R plasmids can provide resistance against particular bacteriophages by encoding specific enzymes.
- Prevalence and Implications:
- R plasmids typically exist in low copy numbers within bacterial cells and can vary in size.
- Their ability to confer multi-drug resistance has profound implications for public health, as highlighted by the potential resistance development in pathogens like the gonorrhea-causing bacterium.
In summation, R plasmids represent a critical aspect of bacterial genetics, facilitating their survival in antibiotic-rich environments. Their role in the rapid emergence of antibiotic-resistant bacterial strains underscores the need for judicious antibiotic use and continued research into combating resistance.
3. Col Plasmids (Colicin plasmids)
Bacteriocinogen, commonly referred to as the Col plasmid, is a specialized genetic element found in certain bacteria, enabling them to produce bacteriocins. Bacteriocins are potent proteins that exhibit bactericidal properties, targeting and eliminating closely related bacterial species or even different strains of the same species. Here are the key features and implications of the Col plasmid:
- Nature of Bacteriocins:
- Bacteriocins, such as colicins, are proteins synthesized by bacteria possessing the Col plasmid. These proteins are lethal to other bacteria, providing a competitive advantage to the producing bacterium.
- Colicins, specifically, are produced by Escherichia coli (E. coli) and are derived from the ColE1 plasmid.
- Bacteriocin Production:
- Bacteriocins are not exclusive to coliforms; various bacteria produce these substances. Collectively, these antibacterial proteins are termed bacteriocins.
- Examples include:
- Colicins: Produced by E. coli.
- Pyocins: Synthesized by Pseudomonas aeruginosa.
- Marscesins: Originating from Serratia marcescens.
- Diphthericin: Produced by Corynebacterium diphtheria.
- Role in Bacterial Defense:
- Bacteriocins play a pivotal role in bacterial defense mechanisms. By producing these lethal proteins, bacteria can eliminate competing strains or species in their vicinity, ensuring their dominance in a particular niche.
- Bacteria lacking the Col plasmid are susceptible to the bactericidal effects of these proteins, highlighting the protective advantage conferred by the Col plasmid.
- Significance in Microbial Typing:
- The production of specific bacteriocins by different bacterial strains aids in the interspecies typing of organisms. This characteristic can be harnessed in microbial research and diagnostics to differentiate between bacterial species or strains.
- Presence in E. coli:
- The Col plasmid, particularly associated with the production of colicins, is frequently observed in E. coli. This bacterium utilizes colicins as a defense mechanism against competing bacterial strains.
In summary, the Bacteriocinogen or Col plasmid represents a fascinating aspect of bacterial genetics, underscoring the intricate defense mechanisms employed by bacteria to thrive in competitive environments. The production of bacteriocins, such as colicins, exemplifies the evolutionary strategies adopted by bacteria to ensure their survival and dominance.
4. Degradative Plasmids
Degradative plasmids play a pivotal role in the metabolic versatility of certain bacteria, enabling them to utilize and degrade compounds that are uncommon in natural environments. Here are the salient features and implications of degradative plasmids:
- Metabolic Capabilities:
- These plasmids are equipped with genes encoding specialized enzymes. These enzymes facilitate the breakdown of specific, often rare, compounds such as toluene, camphor, xylene, and salicylic acid.
- Ecological Significance:
- Degradative plasmids contribute to the ecological balance by assisting in the decomposition of dead organic matter derived from plants and animals. By breaking down these compounds, bacteria harboring degradative plasmids play a role in recycling organic matter, ensuring the continuous availability of essential nutrients in ecosystems.
- Energy Production and Biosynthesis:
- The degradation of uncommon compounds not only aids in recycling but also provides an energy source for the bacterium. The breakdown products can be channeled into various biosynthetic pathways, supporting the growth and reproduction of the bacterial cell.
- Conjugative Nature:
- Degradative plasmids are conjugative, meaning they have the ability to transfer between bacterial cells. This transfer ensures the dissemination of the capability to degrade specific compounds across bacterial populations, enhancing the overall metabolic diversity within microbial communities.
- Chemical Breakdown:
- The presence of degradative plasmids in a bacterial organism empowers it to metabolize a range of chemicals and substances. This metabolic adaptability can be crucial for survival in environments where conventional nutrients are scarce but unusual compounds are available.
In essence, degradative plasmids underscore the remarkable adaptability of bacteria, allowing them to exploit diverse chemical compounds for sustenance. These plasmids not only enhance the metabolic repertoire of the host bacterium but also contribute to the ecological dynamics by facilitating the recycling of organic matter.
5. Virulence Plasmids
Virulence plasmids are specialized genetic elements that confer pathogenic properties to bacteria, enabling them to cause disease in host organisms. Here are the key characteristics and implications of virulence plasmids:
- Pathogenic Transformation:
- The presence of a virulence plasmid within a bacterium can transform an otherwise harmless microorganism into a potent pathogen. This transformation is due to the genes carried by the plasmid, which encode for specific virulence factors.
- Disease Causation:
- Bacteria equipped with virulence plasmids possess the ability to invade host tissues, evade the host’s immune response, and establish infections. As a result, they can cause a range of diseases, from mild to severe, depending on the virulence factors they express.
- Prominent Examples:
- Escherichia coli (E. coli) is a prime example. While many strains of E. coli are benign residents of the human gut, certain strains equipped with virulence plasmids can cause gastrointestinal disturbances, leading to symptoms like severe diarrhea and vomiting.
- Another notable bacterium is Salmonella enterica, which harbors virulence plasmids that contribute to its pathogenicity.
- Disease Transmission:
- Bacteria containing virulence plasmids can be easily transmitted among hosts, leading to outbreaks of infectious diseases. Their ability to rapidly replicate in affected individuals further exacerbates the spread.
- Specific Virulence Factors:
- Virulence plasmids encode for a variety of factors that aid in the bacterium’s pathogenicity. These can include toxins, adhesion molecules, and enzymes that help the bacterium invade host tissues and evade the immune response.
- Agricultural Impact:
- Not only do virulence plasmids impact human health, but they also have implications in agriculture. For instance, the Ti plasmid found in Agrobacterium tumefaciens induces crown gall disease in plants, affecting crop yields.
In summary, virulence plasmids play a pivotal role in the pathogenicity of certain bacteria, enabling them to cause diseases in humans, animals, and plants. Understanding these plasmids is crucial for developing strategies to combat bacterial infections and mitigate their impact on public health and agriculture.
6. Episome
- Episomes are an abacus-like plasmid or viral DNA that has the ability to incorporate itself in the DNA chromosomal an organism that hosts it .
- This is why it is able to remain in place for a lengthy period and be replicated in every cell division in its host. It can also eventually become an essential part of its genetic composition.
Types of Plasmids based on their Ability To Transfer To Other Bacteria
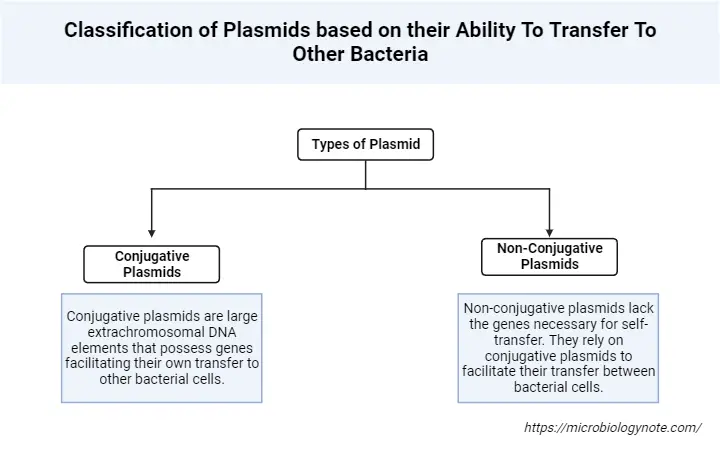
Plasmids, the extrachromosomal DNA elements in bacteria, play a pivotal role in bacterial genetics and physiology. One of the key features of certain plasmids is their ability to transfer between bacterial cells. Based on this ability, plasmids can be broadly classified into two main categories:
- Conjugative Plasmids:
- Definition: Conjugative plasmids are large extrachromosomal DNA elements that possess genes facilitating their own transfer to other bacterial cells.
- Characteristics:
- These plasmids encode genes responsible for the synthesis of sex pili, which are structures that facilitate direct cell-to-cell contact, enabling the transfer of genetic material.
- Conjugative plasmids play a central role in bacterial conjugation, a process akin to sexual reproduction in bacteria, where genetic material is transferred from a donor to a recipient cell.
- Significance: The presence of conjugative plasmids enhances the genetic diversity of bacterial populations, allowing for the rapid spread of advantageous traits, such as antibiotic resistance.
- Non-Conjugative Plasmids:
- Definition: Non-conjugative plasmids lack the genes necessary for self-transfer. They rely on conjugative plasmids to facilitate their transfer between bacterial cells.
- Characteristics:
- Typically found in both Gram-positive and Gram-negative bacteria, including species like Haemophilus influenzae and Neisseria gonorrhoeae.
- These plasmids are generally smaller in size, often ranging from 1-10 daltons.
- A single bacterium can harbor multiple copies of non-conjugative plasmids, sometimes exceeding 30 copies per cell.
- Mobilization: In situations where a bacterium possesses both conjugative and non-conjugative plasmids, the non-conjugative plasmids can be “mobilized” for transfer. This means that while they cannot initiate the transfer process on their own, they can be co-transferred alongside conjugative plasmids during conjugation.
Additional Classification – Incompatibility:
- Plasmids within a bacterium must be compatible to coexist. If two plasmids are incompatible, one will be eliminated from the cell.
- Incompatibility arises when plasmids utilize the same replication or partitioning mechanism within the cell. This ensures that each plasmid can maintain its distinct territory within the bacterial cell without interference from others.
In conclusion, the ability of plasmids to transfer between bacterial cells has profound implications for bacterial evolution, adaptation, and pathogenicity. Understanding the mechanisms and classifications of plasmid transfer is crucial for insights into bacterial genetics and the spread of antibiotic resistance.
Replication of Plasmids
Plasmids, the extrachromosomal DNA elements found in bacteria, possess the unique ability to replicate independently of the bacterial chromosome. This independent replication is facilitated by specific origins of replication present within the plasmid DNA. Here’s an in-depth look into the process:
- Enzymatic Involvement:
- Plasmids utilize common cellular enzymes for their replication, especially in the case of smaller plasmids.
- Some specialized plasmids, however, encode their own specific enzymes that aid in their replication process, ensuring efficient and controlled replication.
- Genetic Control:
- Plasmids typically contain a limited number of genes, often fewer than 30.
- A significant portion of these genes is dedicated to controlling the initiation of replication and ensuring the proper distribution of replicated plasmids to daughter cells during cell division.
- It’s noteworthy that the genetic information within plasmids is not essential for the host bacterium’s survival. Bacteria can function normally even in the absence of these plasmids.
- Speed of Replication:
- Due to their compact size, plasmids can replicate rapidly, often completing the process in a fraction of the time it takes for the entire bacterial cell to divide.
- Mechanisms of Replication:
- Most gram-negative bacteria plasmids replicate using a mechanism akin to the bacterial chromosomal replication. This process, known as bidirectional replication, starts at the origin of replication and progresses in two directions, forming a structure reminiscent of the Greek letter Theta (Θ).
- Some gram-negative bacterial plasmids employ a unidirectional replication method.
- In contrast, many plasmids in gram-positive bacteria adopt a replication strategy similar to the phage φX174, which involves a rolling circle mechanism.
- Linear plasmids, on the other hand, often employ a unique replication mechanism where a protein is attached to the 5′-ends of the DNA. This protein plays a pivotal role in initiating DNA synthesis.
In summary, plasmid replication is a complex yet efficient process that ensures the propagation of these extrachromosomal elements within bacterial populations. Understanding this process provides insights into bacterial genetics, biotechnology applications, and the spread of antibiotic resistance.
Plasmid Vector
Plasmid vectors play a pivotal role in the realm of genetic engineering and biotechnology. These vectors, often derived from plasmids and bacteriophages, serve as vehicles to introduce foreign DNA into host organisms, enabling the expression and study of the introduced genes.
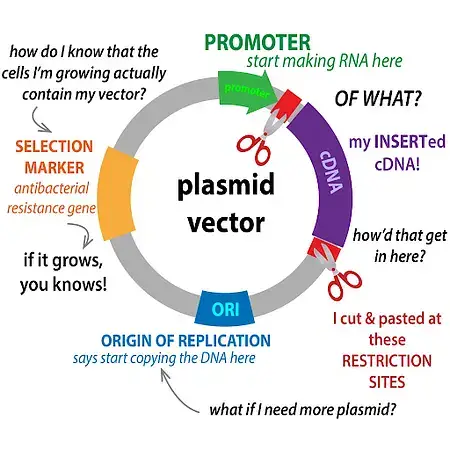
Key Components of Plasmid Vectors:
- Origin of Replication (ORI):
- This is a specific DNA sequence where the replication process commences. It ensures that the plasmid is replicated within the host organism, allowing for the propagation of the introduced gene.
- Selectable Marker:
- These markers, often genes conferring antibiotic resistance, facilitate the identification and selection of host cells that have successfully taken up the plasmid vector. This ensures that only cells containing the desired plasmid are grown and studied.
- Multiple Cloning Sites (MCS):
- These are specific DNA sequences recognized by restriction enzymes. They allow for the precise insertion of foreign DNA fragments into the plasmid vector. Ideally, a single or few recognition sites are preferred to maintain the integrity of the inserted gene.
- Promoter Region:
- This region drives the transcription of the inserted gene, ensuring that the gene is expressed and the corresponding protein is produced within the host organism.
- Primer Binding Site:
- Essential for techniques like PCR amplification, this DNA sequence serves as the starting point for replication and sequencing processes.
Process of Utilizing Plasmid Vectors:
To harness the capabilities of plasmid vectors, specific steps are undertaken:
- DNA of interest is isolated and cut at specific sites using restriction enzymes, producing “sticky ends”.
- The plasmid vector is similarly cut, and the DNA of interest is inserted using DNA ligase, creating a recombinant DNA molecule.
- This recombinant plasmid is then introduced into a host organism, typically a bacterial cell, through a process known as transformation.
- Within the host, the recombinant plasmid replicates, utilizing the host’s DNA polymerase.
- As the host cell divides and grows, the recombinant DNA is also propagated, leading to the production of the desired protein.
Historically, the plasmid pSC101 from Salmonella typhimurium marked a significant milestone as the first plasmid used as a cloning vector. This discovery paved the way for groundbreaking experiments, such as the expression of frog genes within bacterial cells. Today, E. coli plasmids are among the most commonly employed vectors in genetic engineering, testament to their versatility and efficacy.
Isolation of Plasmid
The isolation of plasmid DNA is a fundamental procedure in molecular biology, enabling researchers to obtain pure plasmid DNA for applications such as cloning, transfection, and polymerase chain reaction (PCR). The process is meticulously designed to extract plasmid DNA from host cells, ensuring its integrity and purity. Here’s a detailed overview of the plasmid isolation process:
1. Cell Cultivation:
- The initial step involves cultivating bacteria that harbor the desired plasmid in a specific culture medium. To ensure the selective growth of plasmid-containing bacteria, antibiotics can be added to the medium, suppressing the growth of non-plasmid bearing bacteria.
2. Centrifugation:
- Following bacterial cultivation, the cells are subjected to centrifugation. This step separates the bacterial cells from the culture medium. The resulting pellet contains the bacterial cells, which are the primary source of plasmid DNA.
3. Alkaline Lysis Method:
- One of the most widely adopted techniques for plasmid isolation is the alkaline lysis method, which involves the following steps:
- Resuspension: The bacterial pellet obtained from centrifugation is resuspended in an isotonic solution containing ethylenediaminetetraacetic acid (EDTA). EDTA acts as a chelating agent, inhibiting nuclease activity and ensuring DNA integrity.
- Cell Lysis: The resuspended cells are treated with an alkaline solution containing sodium dodecyl sulfate (SDS). This disrupts the cell membrane, releasing the cellular contents, including plasmid DNA.
- Protein Precipitation: Acidic potassium acetate is added to the lysate, leading to the precipitation of cellular proteins and other impurities.
- Sedimentation: The mixture is centrifuged, allowing the sedimentation of cellular debris and proteins, leaving the plasmid DNA in the supernatant.
4. Purification:
- To further purify the plasmid DNA, a mixture of phenol and chloroform is used. This organic extraction step removes residual proteins and other contaminants from the DNA solution.
5. DNA Precipitation:
- Ethanol is added to the purified DNA solution to precipitate the plasmid DNA. This is followed by centrifugation to collect the DNA pellet.
6. Washing and Resuspension:
- The DNA pellet is washed with 70% ethanol to remove any remaining salts. After a final centrifugation step, the purified plasmid DNA is resuspended in TE buffer (Tris-EDTA) and stored for subsequent applications.
In essence, the isolation of plasmid DNA is a systematic process that ensures the extraction of high-quality, pure plasmid DNA, ready for various molecular biology applications.
Host Range of plasmid
The host range of a plasmid refers to the spectrum of bacterial species in which a particular plasmid can effectively replicate. This characteristic is primarily governed by the origin of replication (ori) region of the plasmid, which dictates the initiation of the replication process.
1. Narrow Host Range Plasmids:
- Characteristics: These plasmids have a limited range of bacterial hosts in which they can replicate.
- Examples: Plasmids of the ColE1 type, such as pBR322, pET, and pUC, fall under this category.
- Host Specificity: Replication of these plasmids is predominantly observed in Escherichia coli. However, they can also replicate in bacteria that are phylogenetically close to E. coli, like Salmonella and Klebsiella.
2. Broad Host Range Plasmids:
- Characteristics: These plasmids possess the ability to replicate in a wide variety of bacterial species, spanning both Gram-negative and Gram-positive bacteria.
- Examples: RK2, RSF1010, and RC plasmids such as pBBR1MCS are representatives of broad host range plasmids.
- Host Specificity:
- Plasmids with the ori region derived from RK2 can replicate in a vast majority of Gram-negative proteobacteria.
- RSF1010-derived plasmids have an even broader host range, with the capability to replicate in Gram-positive bacteria, including those from the Firmicutes group.
- The unique feature of broad host range plasmids is their ability to replicate even in bacterial species that are distantly related phylogenetically.
- Replication Mechanism: One of the distinctive attributes of broad host range plasmids is their relative independence from the host cell’s machinery for replication. This is because they encode their own proteins essential for initiating the replication process.
Applications: Given their versatility, broad host range plasmids are invaluable tools in molecular biology, especially when gene expression is required across diverse bacterial species.
In summary, the host range of a plasmid is a crucial determinant of its utility in genetic engineering and molecular biology. Whether narrow or broad in their host range, plasmids play a pivotal role in advancing our understanding of bacterial genetics and facilitating various biotechnological applications.
Determining the Host Range
Determining the host range of a plasmid is a pivotal aspect of molecular biology, especially when assessing the potential of a plasmid to replicate in different bacterial species. The host range essentially dictates the types of bacteria in which a specific plasmid can effectively replicate.
1. Introduction to Host Range Determination:
- The precise host range of many plasmids remains elusive due to the challenges associated with ascertaining whether a given plasmid can replicate in a specific bacterial host.
2. Transformation:
- This is a foundational method employed to introduce plasmids into bacteria to assess their replication capability.
- However, the transformation process is not universally applicable to all bacterial species, presenting a limitation in its utility.
3. Electroporation:
- An alternative to transformation, electroporation is a technique that facilitates the introduction of DNA, including plasmids, into cells by applying an electric field.
4. Conjugation:
- This process allows for the direct transfer of plasmids between bacterial cells, offering another avenue to introduce plasmids into different bacterial species.
5. Gene Expression and Resistance:
- It is noteworthy that a gene present on a plasmid may not always be expressed optimally in a different bacterial host.
- Certain genes, especially those conferring antibiotic resistance, can be transferred between bacteria. For instance, the kanamycin resistance gene, originally identified in the Tn5 transposon, can confer resistance in a wide range of Gram-negative bacteria.
6. Cloning and Transposons:
- The ability to confer antibiotic resistance can be harnessed to clone marker genes in plasmids, facilitating their amplification.
- Transposons carrying selectable markers can also be integrated into plasmids to enhance their utility.
7. Challenges and Considerations:
- It is imperative to ensure that introduced plasmids do not recombine with the host chromosome, as this can confound results.
- The process of determining the host range is labor-intensive and fraught with challenges. Various barriers can impede the successful transfer of plasmids into host bacteria. Moreover, a single method may not be universally applicable to all plasmids and bacterial species.
In conclusion, while determining the host range of a plasmid is of paramount importance in molecular biology, it is a complex endeavor that requires meticulous planning and execution. The choice of method and the inherent challenges associated with each technique necessitate a comprehensive understanding of both the plasmid in question and the target bacterial species.
Transfer of Plasmid
Plasmid transfer is a fundamental process in bacterial genetics, allowing for the exchange of genetic material between bacterial cells. This transfer predominantly occurs through a mechanism known as conjugation.
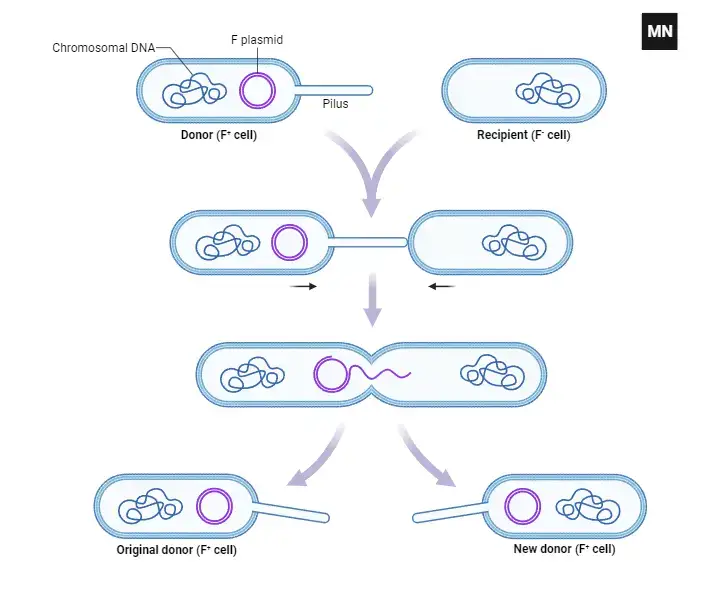
Mechanism of Plasmid Transfer through Conjugation:
- Cellular Participants:
- Conjugation involves two bacterial cells: the donor, which possesses the plasmid, and the recipient, which receives it.
- Formation of the Conjugation Bridge:
- The donor cell synthesizes a specialized structure known as the pilus. This pilus acts as a conjugation bridge, facilitating the direct physical connection between the donor and recipient cells.
- Transfer of the Plasmid:
- Once the cells are connected via the pilus, the donor cell replicates its plasmid and transfers a copy to the recipient cell. This ensures that both cells retain a copy of the plasmid post-conjugation.
- Alternative Transfer Mechanisms:
- While conjugation is a primary mode of plasmid transfer, other mechanisms also exist. Transduction involves the use of bacteriophages (viruses that infect bacteria) to transfer plasmids between bacterial cells.
- Bacterial transformation, on the other hand, is a process where bacteria take up external DNA from their environment, which can include plasmids.
In summary, the transfer of plasmids plays a pivotal role in bacterial genetics, enabling the dissemination of genetic traits across bacterial populations. Conjugation, as a primary mode of transfer, underscores the intricate mechanisms bacteria have evolved to share genetic information.
Functions and Applications of Plasmids
Plasmids, the small, circular DNA molecules found in bacterial cells, play a pivotal role in various biological processes and have been harnessed for numerous applications in biotechnology and medicine. Here’s an overview of their functions and applications:
- Antibiotic Resistance:
- One of the primary roles of plasmids is to confer antibiotic resistance to bacteria. They carry specific resistance genes that can be transferred between bacterial cells, allowing for the spread of resistance within bacterial populations.
- Recombinant DNA Technology:
- Plasmids are integral to recombinant DNA technology. Their ability to incorporate foreign genes and replicate within bacterial cells makes them ideal vectors for genetic engineering.
- Drug Delivery:
- Plasmids have been explored as vehicles for delivering specific drugs to targeted cells in the body, enhancing the efficacy and specificity of treatments.
- Production of Therapeutic Proteins:
- Plasmids have been employed to produce essential human proteins, such as insulin and growth hormones, in mammalian cells. This biotechnological application has revolutionized the treatment of diseases like diabetes.
- Gene Therapy:
- In the realm of gene therapy, plasmids are used to introduce therapeutic genes into the human body to combat various diseases. Their ability to target defective cells and activate therapeutic genes makes them invaluable in this domain.
- Environmental Applications:
- Plasmids play a role in environmental biotechnology. They carry genes that enable bacteria to digest pollutants, aiding in the bioremediation of contaminated environments.
- Antibacterial Protein Production:
- Certain plasmids encode for proteins with antibacterial properties, providing a competitive advantage to the host bacterium.
- Pathogenicity Enhancement:
- Some plasmids carry genes that enhance the virulence or pathogenicity of bacteria, allowing them to cause diseases more effectively.
- Metabolic Adaptations:
- Plasmids can equip bacteria with metabolic advantages, such as the ability to fix nitrogen or degrade specific organic compounds, especially under nutrient-limited conditions.
- Vectors in Genetic Engineering:
- Due to their ease of manipulation and replication capabilities, plasmids serve as vectors in genetic engineering, facilitating the transfer of genes between organisms.
- Horizontal Gene Transfer:
- Plasmids are agents of horizontal gene transfer, allowing for the exchange of genetic material between bacteria, leading to increased genetic diversity.
In conclusion, plasmids are versatile genetic elements with a wide range of functions and applications. Their role in antibiotic resistance, gene therapy, and biotechnology underscores their significance in both natural ecosystems and human-made systems.
Plasmids and Ampicillin Resistance: Certain plasmids contain ampR genes, which confers resistance to ampicillin, an antibiotic. E. bacteria cells that have this plasmid, referred to as “+ampR” cells are able to thrive and grow into colonies on LB Agar which is added with ampicillin. However, cells that do not have ampR plasmids, also known as “-ampR” cells are tolerant to the antibiotic that eliminates them. The ampicillin-sensitive cells ( ampR) is able to transform into the ampicillin-resistant (+ampR) cell through the acceptance of an external plasmid that contains ampR genes.
Advantages of Plasmids
Plasmids, the small, circular DNA molecules independent of the chromosomal DNA in bacteria, offer several advantages, both naturally and in biotechnological applications. Here are the key advantages of plasmids:
- Gene Transfer:
- Plasmids can facilitate horizontal gene transfer between bacteria, allowing for the exchange of genetic information and increasing genetic diversity.
- Antibiotic Resistance:
- Plasmids often carry antibiotic resistance genes, providing bacteria with a survival advantage in environments with antibiotic presence.
- Environmental Adaptability:
- Plasmids can carry genes that allow bacteria to metabolize specific compounds, aiding in survival in varied environments.
- Recombinant DNA Technology:
- Plasmids are widely used as vectors in genetic engineering due to their ability to incorporate foreign DNA and replicate independently.
- Easy Manipulation:
- Plasmids are relatively easy to isolate, modify, and reintroduce into bacterial cells, making them ideal tools for molecular biology research.
- High Copy Number:
- Some plasmids can exist in multiple copies within a single bacterial cell, allowing for the amplification of genes they carry.
- Gene Therapy:
- Plasmids are being explored as vectors for delivering therapeutic genes to treat certain diseases in humans.
- Protein Production:
- Plasmids can be engineered to express proteins in bacterial cells, facilitating large-scale protein production for research, pharmaceuticals, and industrial applications.
- Bioremediation:
- Plasmids can carry genes that enable bacteria to degrade environmental pollutants, making them valuable in bioremediation efforts.
- Vaccine Development:
- Plasmids can be used to develop DNA vaccines by inserting genes that code for antigens into the plasmid and then introducing it into the body to stimulate an immune response.
- Research Tool:
- Plasmids are essential tools in labs for cloning, gene expression studies, and creating genetically modified organisms.
- Cost-Effective:
- Using plasmids for protein production or gene expression in bacteria is often more cost-effective than other eukaryotic expression systems.
- Safety:
- Plasmids, being non-viral vectors, pose fewer safety concerns when used in gene therapy compared to viral vectors.
In summary, plasmids play a crucial role in bacterial adaptability and survival and have been instrumental in advancing genetic and biotechnological research. Their versatility and ease of use make them invaluable tools in various scientific and medical applications.
Disadvantages of Plasmids
While plasmids offer numerous advantages, especially in the realm of biotechnology and genetic engineering, they also come with certain limitations and potential drawbacks. Here are the key disadvantages of plasmids:
- Limited Size of Insert:
- Plasmids can only carry a limited size of foreign DNA insert. Larger genes or sequences may not be easily cloned into standard plasmids.
- Stability Issues:
- Over time, especially under non-selective conditions, bacteria might lose the plasmid, leading to instability in genetic experiments.
- Antibiotic Resistance Spread:
- Plasmids can facilitate the spread of antibiotic resistance genes among bacterial populations, contributing to the global issue of antibiotic resistance.
- Potential for Unintended Transfer:
- There’s a risk of horizontal gene transfer to non-target organisms in the environment, leading to unforeseen ecological consequences.
- Limited Expression in Eukaryotes:
- Plasmids are primarily bacterial vectors, and their use in eukaryotic cells can be challenging due to differences in gene expression mechanisms.
- Safety Concerns in Therapy:
- While plasmids are considered safer than viral vectors, there’s still a risk of unintended genetic changes when used in gene therapy.
- Low Transfection Efficiency:
- In some eukaryotic cells, especially primary cells, the efficiency of plasmid entry (transfection) can be quite low compared to viral vectors.
- Potential for Mutation:
- Replication and manipulation of plasmids can sometimes introduce mutations, which might affect the function of the cloned gene.
- Cost:
- While bacterial systems using plasmids are generally cost-effective, the initial setup, especially for large-scale experiments or production, can be expensive.
- Overexpression Issues:
- Overexpression of certain genes from plasmids can be toxic to host cells, affecting cell health and viability.
- Regulatory and Ethical Concerns:
- The use of plasmids in creating genetically modified organisms (GMOs) can raise regulatory and ethical issues, especially when released into the environment.
- Dependency on Antibiotics:
- Many plasmid systems rely on antibiotics for selection, which can contribute to antibiotic consumption in labs and potential downstream effects.
- Recombination Risks:
- There’s a potential risk of recombination events between plasmids and chromosomal DNA, leading to unintended genetic changes.
While plasmids are invaluable tools in research and biotechnology, it’s essential to be aware of their limitations and potential risks, ensuring their use is both effective and safe.
Example of Plasmids
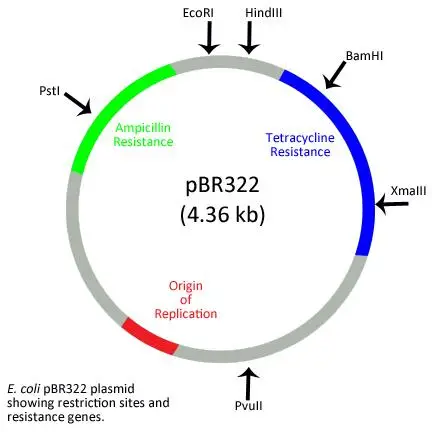
1. pBR322 Plasmid
The pBR322 plasmid stands as a pioneering achievement in the realm of molecular biology, having been crafted as the first artificial cloning vector in 1977 by Bolivar and Rodriguez. The nomenclature “pBR322” is derived from its components: “P” represents Plasmid, “BR” signifies the creators Bolivar and Rodriguez, and “322” is a unique identifier for this specific plasmid among others developed in the same laboratory.
Key Features of pBR322:
- Antibiotic Resistance Genes:
- The plasmid is equipped with two primary resistance genes: Ampicillin Resistance (ampR) and Tetracycline Resistance (tetR).
- These genes serve as selectable markers, facilitating the identification and selection of host cells that have undergone successful transformation with the vector. Concurrently, they aid in the elimination of non-transformed cells.
- Restriction Sites:
- pBR322 boasts a plethora of restriction endonuclease recognition sites, including but not limited to BamH I, Hind III, Sal I, Pvu I, Pvu II, Pst I, EcoR I, and Cla I.
- Origin of Replication (ORI):
- This is a crucial component that dictates the initiation of the plasmid’s replication process.
- ROP Region:
- This segment encodes proteins pivotal for the plasmid’s replication mechanism.
- Insertion of Foreign DNA:
- The antibiotic resistance genes within pBR322 can also function as restriction sites for the insertion of foreign DNA. When foreign DNA is integrated, the corresponding resistance gene becomes inoperative, thus aiding in the differentiation of transformed and non-transformed cells.
- Alternative Selectable Markers:
- Beyond antibiotic resistance, pBR322 can employ alternative markers, such as the gene encoding for β-galactosidase. This enzyme, when present, can react with a chromogenic substrate to produce a distinct color.
- If a foreign gene is inserted within the β-galactosidase gene, the resultant recombinant cell ceases to produce the enzyme due to gene inactivation. Consequently, in the presence of a chromogenic substrate, non-recombinant cells yield blue colonies, while recombinant cells produce colorless colonies.
In summary, the pBR322 plasmid is a foundational tool in genetic engineering, offering a versatile platform for cloning and transformation experiments. Its design, inclusive of multiple resistance genes and restriction sites, ensures precision and efficiency in molecular biology research.
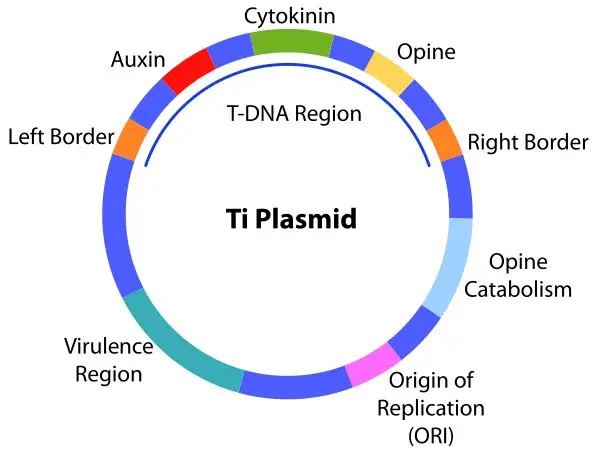
2. Ti Plasmid
The Ti Plasmid, an abbreviation for tumor-inducing plasmid, is a distinctive genetic element found within the bacterium Agrobacterium tumifaciens. Renowned for its role in plant pathology, this plasmid has been harnessed for its unique capability to transfer genes into plant cells, earning Agrobacterium tumifaciens the moniker “nature’s genetic engineer.”
Key Attributes of the Ti Plasmid:
- Size and Structure:
- The Ti plasmid is substantial, with an approximate size of 250 kbp.
- It encompasses one or more T-DNA regions, which are essential for its gene transfer capability.
- Function and Pathogenicity:
- The bacterium Agrobacterium tumifaciens, armed with the Ti plasmid, induces crown gall disease in a myriad of dicotyledonous plants.
- The plasmid’s T-DNA segment is transferred into plant cells, instigating a transformation that prompts these cells to form tumors. Concurrently, these transformed cells synthesize specific chemicals that benefit the bacterium.
- Genetic Diversity:
- Various Ti plasmids exist, each characterized by distinct genes that code for specific opines such as leucinopine, nopaline, and octopine.
- Genetic Engineering Potential:
- The Ti plasmid’s inherent ability to transfer genes has been exploited for biotechnological applications. By introducing desired genes into the Ti plasmid, researchers can ensure their subsequent integration into plant cells.
- Notably, once the desired gene is integrated, the Ti plasmid loses its pathogenic traits, ensuring that while the desired gene is incorporated, the plant remains free from disease.
- Virulence Genes:
- The Ti plasmid houses virulence genes, abbreviated as “vir.” These genes are instrumental in transferring the T-DNA region into plant cells, where it integrates with the plant genome.
- Modifiability:
- One of the Ti plasmid’s most advantageous features is its adaptability. Researchers can modify it to accommodate specific genes, tailoring it for various genetic engineering endeavors.
In essence, the Ti plasmid stands as a testament to nature’s ingenuity, offering a natural mechanism for gene transfer. Its attributes have been pivotal in advancing plant biotechnology, allowing for the creation of transgenic plants tailored for specific needs.
Quiz Practice
What is a plasmid?
a) A type of virus
b) A small, circular piece of DNA found in bacteria
c) A protein structure in bacterial cells
d) A type of bacterial cell wall
Which of the following is NOT a function of plasmids?
a) Antibiotic resistance
b) Photosynthesis
c) Production of toxins
d) Transfer of genes between bacteria
How are plasmids typically transferred between bacterial cells?
a) Transcription
b) Replication
c) Conjugation
d) Translation
Which of the following is a common use of plasmids in genetic engineering?
a) As a cloning vector
b) To increase bacterial growth rate
c) To produce bacterial spores
d) As a source of energy for bacteria
What feature in plasmids allows for the selection of transformed bacteria?
a) Origin of replication
b) Restriction sites
c) Antibiotic resistance genes
d) Pilus genes
Which enzyme is used to cut DNA at specific sites to insert into plasmids?
a) DNA ligase
b) DNA polymerase
c) Restriction endonuclease
d) RNA polymerase
Which of the following plasmids induces tumor formation in plants?
a) pBR322
b) Ti plasmid
c) F plasmid
d) R plasmid
What is the primary purpose of the origin of replication in a plasmid?
a) To provide antibiotic resistance
b) To initiate DNA replication
c) To produce proteins
d) To transfer genes
Which type of plasmid is involved in bacterial conjugation?
a) F plasmid
b) R plasmid
c) Ti plasmid
d) pBR322
Plasmids that exist in multiple copies within a bacterial cell are known as:
a) Conjugative plasmids
b) Low-copy-number plasmids
c) High-copy-number plasmids
d) Transposons
FAQ
What are plasmids?
Plasmids are small, circular DNA molecules that exist separately from the chromosomal DNA in cells. They can carry extra genetic information and replicate independently.
What is the role of plasmids in bacteria?
Plasmids provide bacteria with additional genetic traits, such as antibiotic resistance, virulence factors, or metabolic functions, which can enhance their survival and adaptability in different environments.
How do plasmids replicate?
Plasmids replicate by utilizing specific origin of replication (OR) sites and replication proteins. They can replicate independently within the bacterial cell, producing multiple copies of themselves.
Are plasmids present in all organisms?
Plasmids are commonly found in bacteria, archaea, and some eukaryotic cells. However, their presence and characteristics can vary across different organisms.
Can plasmids transfer genetic information between bacteria?
Yes, plasmids are known for their ability to undergo horizontal gene transfer, transferring genetic information between bacteria. This mechanism contributes to the spread of traits, including antibiotic resistance, among bacterial populations.
Can plasmids be artificially manipulated in the laboratory?
Yes, plasmids are extensively used in genetic engineering and biotechnology. They can be manipulated, modified, and engineered to carry specific genes of interest, making them powerful tools for various applications.
How are plasmids used in molecular biology research?
Plasmids serve as vectors for cloning genes, expressing proteins, and studying gene function. They enable the introduction and manipulation of specific DNA sequences in the laboratory.
What is the copy number of plasmids?
The copy number refers to the number of plasmid copies present within a single bacterial cell. It can vary depending on the size of the plasmid and regulatory factors.
Can plasmids be lost from bacterial cells?
Plasmids can be lost from bacterial cells over time if they provide no selective advantage or if they are not properly segregated during cell division. However, some plasmids have mechanisms to ensure their replicative stability.
Are plasmids only found in pathogenic bacteria?
No, plasmids are found in both pathogenic and non-pathogenic bacteria. While some plasmids carry virulence factors in pathogenic bacteria, many plasmids in non-pathogenic bacteria provide advantageous traits or have unknown functions.
References
- Clark, D. P., Pazdernik, N. J., & McGehee, M. R. (2019). Plasmids. Molecular Biology, 712–748. doi:10.1016/b978-0-12-813288-3.00023-9
- Drlica, K., & Gennaro, M. L. (2001). Plasmids. Encyclopedia of Genetics, 1485–1490. doi:10.1006/rwgn.2001.1000
- Chattoraj, D. K. (2013). Plasmid Replication. Brenner’s Encyclopedia of Genetics, 358–361. doi:10.1016/b978-0-12-374984-0.01173-6
- Filutowicz, M. (2009). Plasmids, Bacterial. Encyclopedia of Microbiology, 644–665. doi:10.1016/b978-012373944-5.00014-6
- https://www.genscript.com/custom-plasmid-preparation.html
- https://royalsocietypublishing.org/doi/10.1098/rstb.2020.0461
- https://www.slideshare.net/Dilippandya/plasmid
- https://www.slideshare.net/doctorrao/plasmids-6828679
- https://www.cd-genomics.com/blog/plasmid-fact-sheet-definition-structure-and-application/
- https://www.news-medical.net/life-sciences/What-are-Plasmids.aspx
- https://www.microscopemaster.com/plasmids.html
- https://www.slideshare.net/15paratr/plasmids-13031139
- https://blog.addgene.org/plasmids-101-what-is-a-plasmid