What is Agrobacterium?
- Agrobacterium is a genus of Gram-negative bacteria developed by H. J. Conn that causes tumors in plants through horizontal gene transfer. Agrobacterium tumefaciens is the species investigated most frequently within this genus.
- Agrobacterium is well-known for its ability to transfer DNA between itself and plants; consequently, it has become an indispensable tool in genetic engineering.
- Prior to the 1990s, Agrobacterium was considered a wastebasket taxon. Many Agrobacterium species, particularly marine species, were reassigned to genera such as Ahrensia, Pseudorhodobacter, Ruegeria, and Stappia with the advent of 16S sequencing.
- The remaining Agrobacterium species have been classified into three biovars: biovar 1 (Agrobacterium tumefaciens), biovar 2 (Agrobacterium rhizogenes), and biovar 3 (Agrobacterium sphaericus) (Agrobacterium vitis). Early in the twenty-first century, Agrobacterium became synonymous with Rhizobium. This action generated controversy.
- The controversy was settled when the genus Agrobacterium was reinstated when it was shown to be phylogenetically separate from Rhizobium. Agrobacterium species are united by a unique synapomorphy: the presence of the protelomerase gene, telA, which confers a linear chromosome on all members of the genus.
- Biovar 1 remained with Agrobacterium, while biovars 2 and 3 were renamed as Rhizobium rhizogenes and Allorhizobium vitis, respectively.
- Agrobacterium tumefaciens is the causative agent of crown gall in plants. The disease is distinguished by a tumor-like growth or gall on the affected plant, frequently near the root-shoot junction.
- The conjugative transfer of a DNA segment (T-DNA) from the bacterial tumour-inducing (Ti) plasmid induces tumors. Agrobacterium rhizogenes, a closely related species, generates root tumors and bears the unique Ri (root-inducing) plasmid.
- Agrobacterium tumefaciens, Agrobacterium rhizogenes, and Agrobacterium vitis are the three biovars found within the genus, despite the fact that the taxonomy of Agrobacterium is now undergoing revision.
- Strains of Agrobacterium tumefaciens and Agrobacterium rhizogenes are known to be able to harbor either a Ti or a Ri-plasmid, whereas strains of Agrobacterium vitis, which are often confined to grapevines, can harbor a Ti-plasmid.
- Non-Agrobacterium strains that harbor a Ri-plasmid have been obtained from environmental samples, and laboratory experiments have demonstrated that non-Agrobacterium strains can also harbor a Ti-plasmid. Certain environmental Agrobacterium strains lack both the Ti and Ri plasmids. These strains are not pathogenic.
- The plasmid T-DNA is semi-randomly integrated into the host cell’s genome, and the tumor morphology genes on the T-DNA are expressed, resulting in the creation of a gall. The T-DNA contains genes for the biosynthetic enzymes responsible for the creation of uncommon amino acids, typically octopine and nopaline.
- It also carries genes for the biosynthesis of the plant hormones auxin and cytokinins, as well as for the biosynthesis of opines, providing a carbon and nitrogen source for the bacteria that most other microorganisms cannot use, giving Agrobacterium a selective advantage.
- By altering the hormone balance in the plant cell, the plant is unable to control the division of those cells, and tumors form. The form of a tumor is determined by the ratio of auxin to cytokinin produced by its genes (root-like, disorganized or shoot-like).
Agrobacterium-mediated Gene Transfer
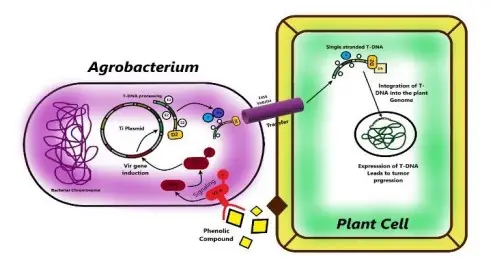
- Agrobacterium-mediated gene transfer, also known as genetic transformation, is a method used to introduce new genes into the DNA of plants. This technique involves the use of a naturally occurring soil bacterium called Agrobacterium tumefaciens, which has the ability to transfer a piece of its own DNA, called the T-DNA, into the host plant’s genome.
- The process of Agrobacterium-mediated gene transfer begins by inserting the desired gene(s) into a plasmid vector, which is a small circular piece of DNA that can replicate independently of the host genome. The plasmid vector also contains the necessary regulatory sequences to control the expression of the inserted gene(s).
- Next, the plasmid vector is introduced into the Agrobacterium tumefaciens through a process called transformation. Once inside the bacterium, the plasmid vector is recognized by the bacterium’s natural transfer machinery, which packages the T-DNA and delivers it into the host plant’s genome.
- The T-DNA is integrated into the host plant’s DNA, resulting in the transfer of the desired gene(s) into the plant’s genome. The transformed plant cells can then be selected and regenerated into whole plants using tissue culture techniques.
- Agrobacterium-mediated gene transfer is a powerful tool for plant genetic engineering, as it allows for the precise and targeted introduction of new genes into plant genomes. This technique has been used to develop crops with improved traits, such as disease resistance, herbicide tolerance, and increased yield.
Factors affecting Agrobacterium-mediated Gene Transfer
Agrobacterium-mediated gene transfer is a widely used technique for introducing foreign DNA into plants. However, successful gene transfer depends on several factors, including:
- Agrobacterium strain: Different strains of Agrobacterium have varying abilities to transfer genes into plants. Therefore, the choice of the strain used is crucial.
- Vector design: The vector used to transfer the gene must be designed in such a way that it can integrate into the plant genome and express the desired gene. The vector also needs to have appropriate regulatory elements that control the expression of the gene.
- Plant species and tissue type: Different plant species and tissue types vary in their susceptibility to Agrobacterium-mediated gene transfer. The choice of the plant species and tissue type should be carefully considered.
- Wounding: Wounding of the plant tissue can enhance gene transfer efficiency. Therefore, some protocols include wounding the plant tissue before Agrobacterium infection.
- Co-cultivation conditions: The co-cultivation conditions, such as temperature, humidity, and light, can also affect gene transfer efficiency. These conditions should be optimized for each specific plant species and tissue type.
- Antibiotics: Antibiotics are often used to select for transformed plant cells. However, the concentration of antibiotics and the duration of selection can also affect gene transfer efficiency.
- Contamination: Contamination with other microorganisms can also reduce gene transfer efficiency. Therefore, it is crucial to maintain sterile conditions throughout the entire process.
- Explants: The type and size of the plant tissue used as an explant can influence gene transfer efficiency. Generally, young and actively growing tissues are more amenable to transformation than mature or differentiated tissues.
- Plant Growth Regulators (PGR): The use of PGRs such as auxins and cytokinins can affect the efficiency of gene transfer. For instance, the addition of cytokinins to the culture medium can enhance shoot regeneration, which can increase the number of transformed plants.
- Light: Light is an important factor that affects plant growth and development. The quality and quantity of light can influence the efficiency of gene transfer. For example, the use of blue light can enhance Agrobacterium-mediated gene transfer in some plant species.
- Temperature: The temperature of the culture environment can also affect gene transfer efficiency. In general, the optimal temperature range for Agrobacterium-mediated gene transfer is between 20-25°C. Higher or lower temperatures can decrease the efficiency of gene transfer.
Why transform plants using Agrobacterium?
Agrobacterium-mediated gene transfer is a preferred method for transforming plants for several reasons:
- High transformation efficiency: Agrobacterium-mediated gene transfer can result in high transformation efficiencies, meaning that a large number of plants can be efficiently transformed with the desired gene(s).
- Precise and targeted gene insertion: Agrobacterium-mediated gene transfer allows for the precise and targeted insertion of new genes into specific sites in the plant genome, resulting in a more stable and predictable gene expression.
- Versatility: Agrobacterium-mediated gene transfer can be used to introduce a wide range of genes into plant genomes, including genes from other species, synthetic genes, and regulatory elements.
- Safe and natural process: Agrobacterium-mediated gene transfer is a natural process that occurs in nature, and does not require the use of harsh chemicals or physical methods that can damage plant cells.
- Compatibility with tissue culture techniques: Agrobacterium-mediated gene transfer is compatible with tissue culture techniques, which allows for the regeneration of whole plants from transformed cells.
Overall, Agrobacterium-mediated gene transfer is a reliable and effective method for introducing new genes into plant genomes, which has led to the development of genetically modified crops with improved traits and increased yields.
Principle of Agrobacterium-mediated Gene Transfer
- The Agrobacterium Ti plasmid’s vir region encodes the majority of the virulence (Vir) proteins utilized by the bacterium to manufacture its T-DNA and deliver it into the plant cell. The T-DNA area (marked by two 25 base pair direct repeats called left and right T-DNA boundaries) is positioned in cis to the vir region on a single Ti plasmid in wild-type Agrobacterium strains.
- In Agrobacterium strains in which the native T-DNA region has been deleted from the Ti plasmid, a recombinant T-DNA region often sits on a tiny, independent binary plasmid and functions trans to the vir region.
- The transformation procedure begins with the adhesion of the bacterium to the plant (step 1), followed by the induction of the vir region’s expression by appropriate host signals (steps 2 and 3). The joint activity of the bacterial VirD1 and VirD2 proteins generates a single-stranded (ss) T-DNA molecule (T-strand) (step 4).
- T-DNA occurs in bacterial cells as a ssDNA–protein complex (immature T-complex) with one VirD2 molecule covalently bound to the 50 end of the T-strand. This complex, along with a number of other Vir proteins, is exported into the host cell (step 5) by a VirB/D4 type IV secretion system, a phase that involves contact between the bacterial Tpilus and at least one host-specific protein.
- T-DNA is believed to reside as a mature T-complex (T-complex) within the cytoplasm of the host cell, where the full length of the T-strand molecule is covered with many VirE2 molecules. These molecules provide the T-DNA with the necessary structure and protection for its journey (step 6) to the nucleus of the host cell.
- The Agrobacterium primarily employs various cellular mechanisms to accomplish the genetic transformation of its host during the final steps of the transformation process, namely transport through the cytoplasm (step 6), nuclear import (step 7), intranuclear transport (step 8), T-DNA uncoating (step 9) and integration (step 10).
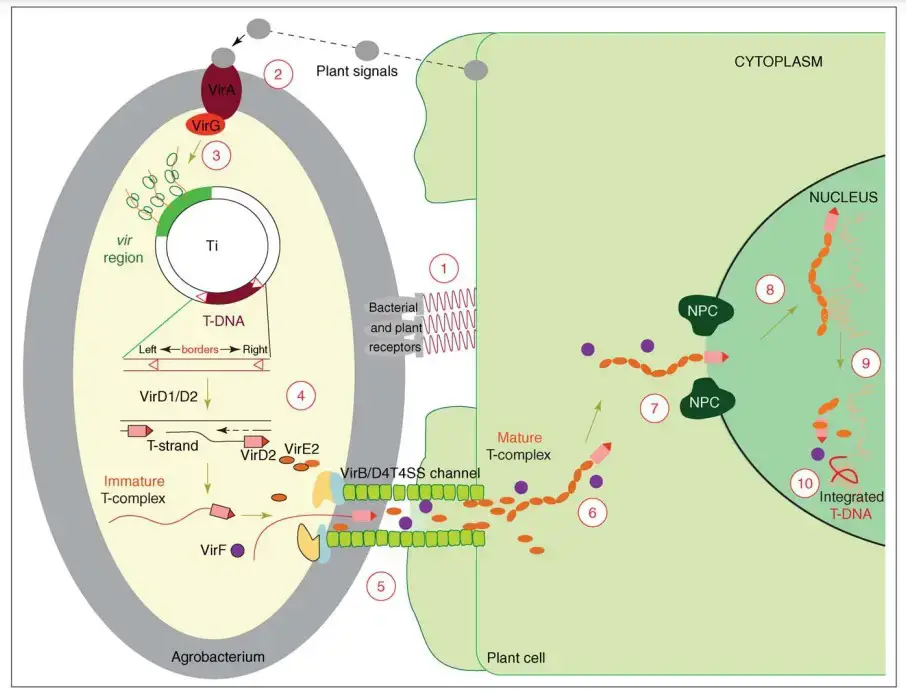
Agrobacterium-mediated transformation mechanism (Natural pathogenesis of A. tumefaciens)
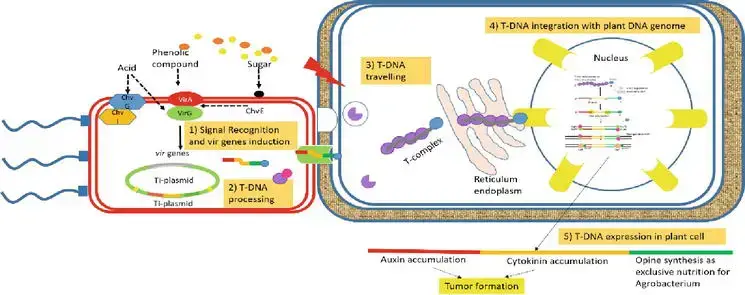
AMT is a complicated mechanism, which includes
- signal recognition from plant host to A. tumefaciens,
- T-DNA processing,
- T-DNA traveling in plant host cell,
- T-DNA integrating to plant host genome, and
- expression of T-DNA in the plant host cell.
The mechanism of T-DNA transfer is facilitated by a set of virulent genes located on Ti-plasmid borne [approximately 35 virulent genes grouped in at least 8 operons, virA, virB, virC, virD, virE, virF, virG, and virH, encoding VirA, VirB, VirC, VirD, VirE, VirF, VirG, and VirH protein, respectively ], apart from T-DNA, whereas others are on chromosome (chromosomal virulent genes—chv).
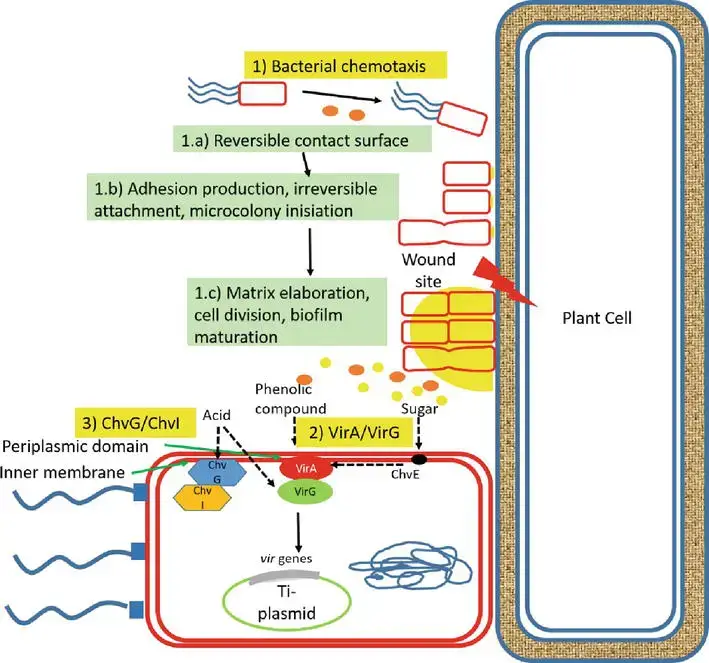
1. Signal recognition
- Large plant-derived chemicals, including organic acid compounds (pH 5.0–5.8) as routinely released chemicals and phenolic compounds as wound-releasing chemicals, interact with Agrobacterium when exposed to the bacteria.
- Three systems are involved in the recognition of A. tumefaciens by plant cells. First, phenolic chemicals, such as acetosyringone and -hydroxy acetosyringone, released from fresh wound sites of plants promote bacterial chemotaxis.
- Initially, bacteria interact with the plant cell surface in a reversible manner, encouraging adhesion synthesis that results in adhesion via unipolar polysaccharide (UPP)-dependent polar attachment and UPP-independent attachment.
- This irreversible surface attachment establishes a site for the creation of multicellular biofilms, matrix elaboration, cell division, biofilm maturation, and the dispersal of “buddy daughter” cells.
- Second, host signal molecules are also detected by transmembrane protein receptors (VirA) in the periplasmic region of bacterial cells, which activate the phosphorylation of the positive regulatory protein VirG. Thirdly, the chromosomal virulent gene-encoded protein ChvG/ChvI detects acidity from sugar, which also activates the basal production of virG.
- The phosphorylation of VirG activates an additional virulence gene. Monosaccharides augment the signaling mechanism by attaching to ChvE and then collaborating with VirA.
2. T-DNA processing
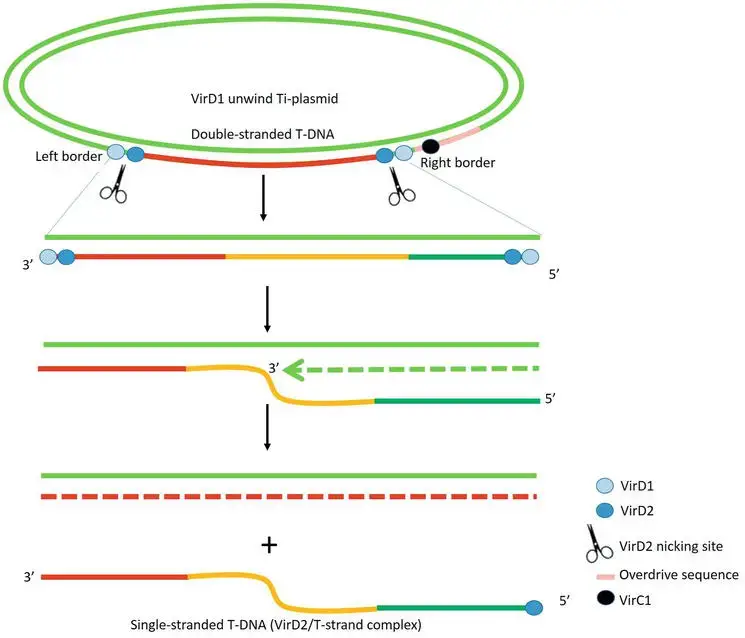
- The process of removing T-DNA from Ti-plasmids is dependent on the endonucleases VirD1 and VirD2. The 25 base pair border sequences on the bottom strand of T-DNA serve as nicking sites for VirD1 and VirD2. Site-specific VirD1 helicase unwinds double-stranded T-DNA.
- The nuclease VirD2 breaks the bottom strand of T-DNA from the right and left border, resulting in T-strand single-stranded linear DNA.
- VirD2 then covalently caps the 5′ end of T-strand at the right border, producing the complex VirD2/T-strand. The 3′ end of the nicked right border serves as a priming point for the regeneration of the bottom strand of T-DNA.
- VirC1 increases the number of T-strand molecules by binding the “overdrive” sequence near the right border of T-DNA via its C-terminal ribbon-helix-helix DNA binding fold.
- The appropriate proportion of the 25 kb terminal sequence of T-DNA determines the DNA transfer director.
3. T-DNA traveling
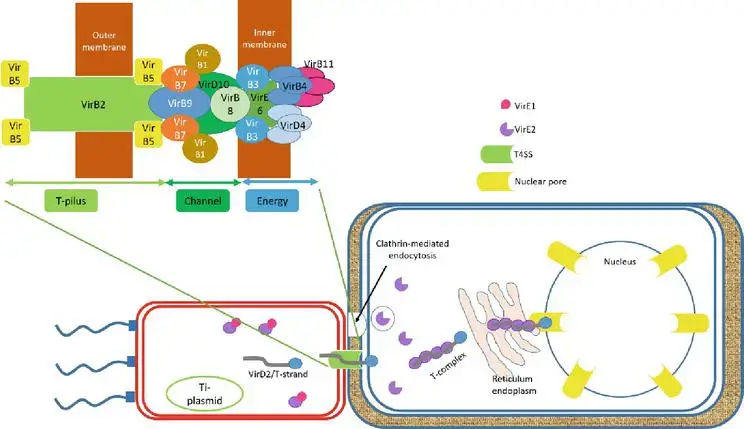
- The role of VirD2 and VirE2 cannot be separated from the T-DNA migrating to the host. Both VirD2 and VirE2 possess the C-terminal nuclear localization signal (NLS) sequence that guided VirD2/T-strand to the nucleus of the host cell.
- VirD2/T-strand, a rod-like structure, exits bacterial cells via the Ti-pilus, type IV secretion system (T4SS), which is composed of 11 VirB and VirD4 proteins. By clathrin-mediated endocytosis, the hydrophilic protein VirE2 accumulates in the bacterial cytoplasm and is translocated into the host cell.
- In addition to aiding in the transport of the T-strand, VirE2 in the host cytoplasm forms the noncovalent VirD2/T-strand/VirE2 (Ti-complex) complex to shield the T-strand from nuclease digestion. Ti-complex is transported to the nucleus of the plant via the endoplasmic reticulum network in the plant cytoplasm.
4. T-DNA integration
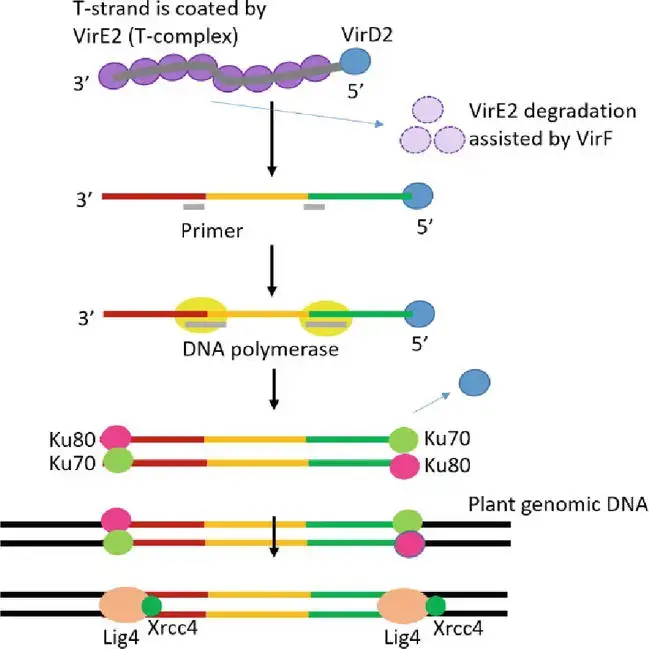
- T-DNA integration followed by transgene expression is the final and most essential step in Agrobacterium-mediated genetic transformation. In actuality, the molecular mechanism of T-DNA integration into the genome of the host plant is not yet entirely understood.
- T-DNA integration occurs at random places, not preferentially in sections of the plant genome that are transcriptionally active or hypomethylated. Certain essential genes for the T-DNA integration process are restricted to those involved in chromatin formation and histone modification.
- Through binding to the bZIP transcription factor VirE2-interacting protein 1, VirE2 may be involved in T-strand localization to chromatin (VIP1).
- VIP1 mediates the connection between VirE2/single-strand DNA and mononucleosomes. As the T-complex arrives in the plant nucleus, its protein component should be destroyed by the ubiquitin-proteasome system in order to reveal the T-strand. VirF facilitates disassembly of the T-complex and degradation of VirE2.
- As VirD2 lacks ligation activity, it is unlikely that T-strands will link directly with the host genome. It is possible that the host DNA polymerase duplicates the T-strand to form a double-stranded T-DNA, which then fuses with site breaks in the DNA of the host plant caused by environmental stress or regular metabolic activities.
- Non-homologous end joining (NHEJ) as a method of repairing broken double-stranded DNA has been hypothesized as the primary process of bacterial plant DNA integration. NHEJ requires minimal or no sequence homology on the damaged region, despite the fact that there is microhomology between the T-DNA and the integration locations on the host chromosome.
5. T-DNA expression
- There are two probable outcomes for bacterial T-DNA that integrates with plant cells. Initially, the T-DNA is expressed at multiple levels. Second, the T-DNA cannot be expressed because it is merely integrated. Depending on the species, transgenic expression might range from very high to completely silent.
- The buildup of auxin and cytokinin is caused by the expression of auxin and cytokinin coding genes in T-DNA. Abnormal ratios of phytohormones cause plant cells to proliferate uncontrollably, resulting to tumor development.
- The expression of opine synthesis coding genes results in the production of opine, the kind of which varies on the bacterial strain and is an exclusive source of nourishment for Agrobacterium.
- In immature tissue, swelling is detected on the fourth or fifth day after bacterial inoculation, is well-developed by one month, and grows rapidly for several months until it reaches two inches in diameter.
Agrobacterium-mediated transformation protocol
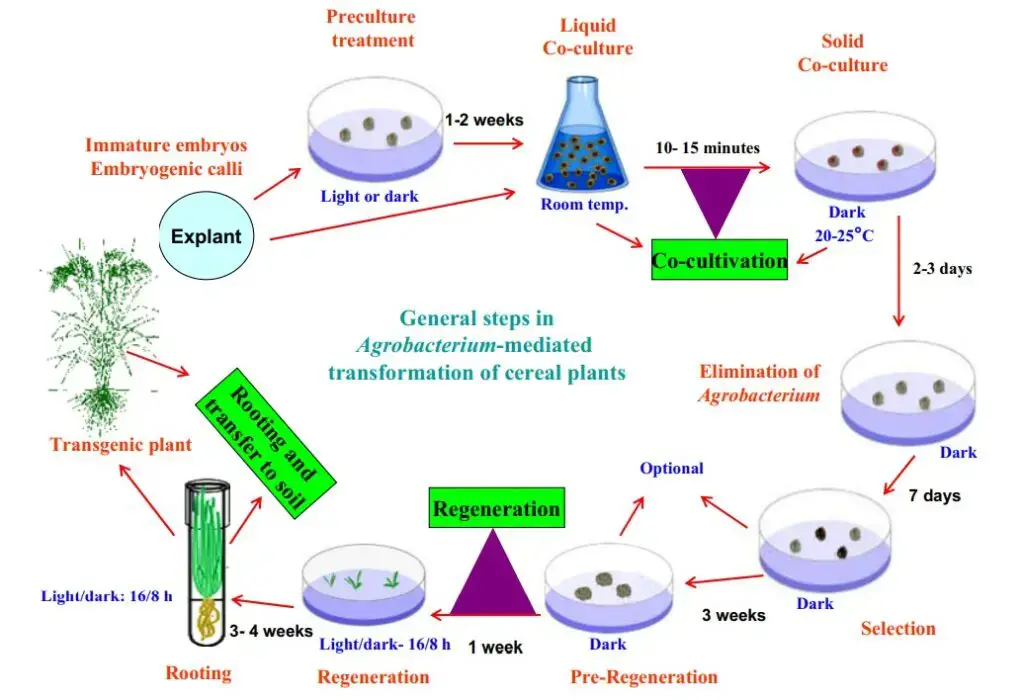
AMT is a generic technique for genetically modifying numerous plant species. Because it facilitates the efficient insertion of stable, non-rearranged, single-copy sequences into the plant genome. The utilization of actively proliferating embryonic callus cells generated from the scutella of mature seeds as the starting material and the addition of a phenolic chemical, acetosyringone, in the cocultivation phases, were identified as crucial for effective transformation. In addition, Cheng et al. revealed that the transformation efficiencies of immature embryos, pre-cultured embryos, and embryogenic callus do not differ significantly.
Various protocols of AMT in either Monocotyledoneae or Dicotyledoneae plants have been documented. Typically, Agrobacterium-based techniques were utilized to create transgenic plants. The procedure consists of seven steps, which are summarized shortly below. (I) preparation of sterile seed or samples and inoculum; (II) explant preparation, infection, and cocultivation with A. tumefaciens; (III) selection; (IV) regeneration; (V) acclimation and molecular identification of T0; (VI) cultivation and self-crossing of T0; and (VII) T1 plant analysis.
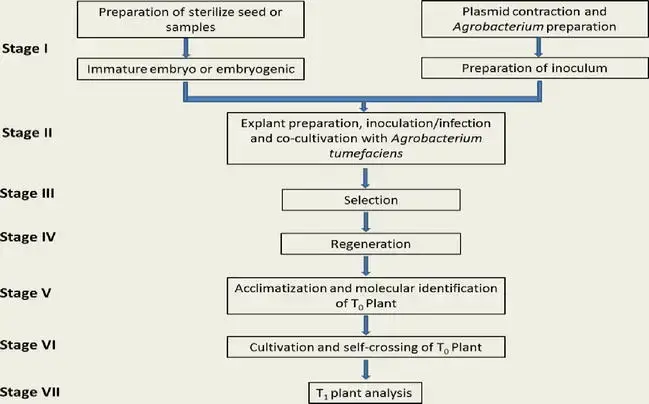
1. Preparation of sterilize seed or samples and inoculum
- Often, embryos were utilized as a sample for transformation. Other research indicates plant transformation efficiencies are genotype or variety dependent.
- To obtain the immature embryo, seeds are planted in sterile media (such as husk, compost, mixed soil, etc.) and cultivated in growth rooms with controlled climatic conditions.
- Depending on the species, immature embryos are harvested following pollination. In contrast, callus can also be generated from hypocotyl or cotyledon explants.
- Inoculum is produced by cultivating an A. tumefaciens strain containing the required antibiotics. The bacteria are cultured in a loop and suspended in media such as LB and LS-inf-AS.
- Inoculum should be freshly prepared. In some instances, it is not required to cultivate A. tumefaciens in liquid culture prior to transformation.
- In contrast, the A. tumefaciens preparation and plasmid creation can be performed in the same manner as the commercial plasmid and A. tumefaciens preparation.
2. Explant preparation, infection, and cocultivation with A. tumefaciens
- As explants, the embryonic, immature embryo or callus can be employed. Before the infection or transformation procedure, the explant should be sterilized. Both the embryo suspension and the bacteria suspension are transferred to the new plate or petri dish.
- After sealing the petri dish, the cocultivation step entails incubating for 2–7 days in the dark at 24–29°C, depending on the species.
- The A. tumefaciens concentration and infection time were discovered to be crucial for preventing explant necrosis and enhancing transformation efficiency. We can adhere to certain protocol recommendations for specific species.
3. Selection
- Selection is one of the most important aspects of transformation’s success. The process of selection can occur after the transformation and regeneration stages, as well as on T0 and T1 plants.
- In addition, antibiotic selection is one of the strategies used to confirm the transformation’s success. In addition to antibiotic selection, PCR should be performed to validate the presence of the transgene of interest in each transformant in each generation.
4. Regeneration
- After the proliferation, the transformed plants underwent regeneration. The branches that have developed from the proliferation explants are removed and planted in a new medium.
- In general, the regeneration step adheres to the in vitro propagation techniques that are separated into shoot regeneration and selection, cut and recut shoot regeneration, and root regeneration.
5. Acclimatization and molecular identification of T0
- After T0’s roots have become robust, acclimatization can take place. The transgenic T0 plant can be cultivated directly in soil or mixed media in a controlled environment or greenhouse.
6. Cultivation and self-crossing of T0
- The primary transformant (T0) was acquired via transformation by A. tumefaciens. After studying the inheritance of transgenes in consecutive generations, T1 seeds are created by self-pollinating the primary transformant (T0). The cultivation process was place in the greenhouse. From the T0 plant, the seed of the primary transformant (T0) is extracted.
7. T1 plant analysis
- T1 plant is the plant produced from the seeds extracted from T0 plant. The examination of T1 plants is concerned with the morphological or physiological expression of the inserted gene.
Applications of Agrobacterium-mediated Gene Transfer
Agrobacterium-mediated gene transfer is a widely used technique with various applications in plant biotechnology. Some of the most common applications include:
- Crop improvement: Agrobacterium-mediated gene transfer can be used to introduce desirable traits into crops, such as disease resistance, herbicide tolerance, and improved yield.
- Functional genomics: The technique can be used to study the function of genes by introducing gene constructs that alter their expression or function.
- Molecular farming: Agrobacterium-mediated gene transfer can be used to produce recombinant proteins in plants. This approach can be more cost-effective and environmentally friendly than traditional methods of protein production.
- Conservation of rare and endangered species: The technique can be used to preserve the genetic diversity of rare and endangered plant species by storing their DNA in a seed bank or by regenerating plants from tissue culture.
- Biosynthesis of novel compounds: Agrobacterium-mediated gene transfer can be used to introduce biosynthetic pathways for the production of novel compounds, such as pharmaceuticals or industrial chemicals, into plants.
- Plant-microbe interactions: The technique can be used to study plant-microbe interactions, such as the role of plant defense genes in response to pathogenic bacteria.
Limitations of Agrobacterium-mediated Gene Transfer
- Limited host range: Agrobacterium-mediated gene transfer is not effective in all plant species. Some plant species are inherently resistant to Agrobacterium infection, and it can be challenging to develop transformation protocols for them.
- Insertion site variability: The site where the transferred DNA integrates into the plant genome is variable and unpredictable. This can lead to positional effects on gene expression, gene silencing, and other unintended effects.
- Tissue culture requirements: The process of Agrobacterium-mediated gene transfer often requires tissue culture, which can be technically challenging, time-consuming, and expensive.
- Epigenetic effects: The transferred DNA can sometimes induce epigenetic changes, such as DNA methylation, that can affect the expression of endogenous genes and the stability of the transgene.
- Unintended effects: The introduction of foreign DNA into plants can sometimes have unintended effects, such as affecting plant development or altering metabolic pathways.
- Regulatory hurdles: The use of Agrobacterium-mediated gene transfer in agriculture is subject to regulatory approval in many countries. This can make it difficult and expensive to commercialize genetically modified plants.
Advantages of Agrobacterium-mediated Gene Transfer
- High transformation efficiency: Agrobacterium-mediated gene transfer can achieve high transformation efficiency, with many transformed plants produced from a single transformation experiment.
- Precise integration of DNA: The transferred DNA integrates into the plant genome at a precise location, which can reduce the likelihood of disrupting endogenous genes or regulatory elements.
- Stability of transgenes: Transgenes introduced via Agrobacterium-mediated gene transfer are often stably inherited and can be passed on to subsequent generations.
- Low risk of pathogenicity: Agrobacterium tumefaciens, the bacterium used in Agrobacterium-mediated gene transfer, is generally considered safe and has a low risk of pathogenicity.
- Easy to use: The technique is relatively easy to use and does not require sophisticated equipment or expertise.
- Large DNA fragments can be transferred: Large DNA fragments, including entire genes and gene clusters, can be transferred using Agrobacterium-mediated gene transfer.
FAQ
What is Agrobacterium-mediated gene transfer?
Agrobacterium-mediated gene transfer is a genetic engineering technique used to transfer foreign genes into the genome of a plant cell using a bacterium called Agrobacterium tumefaciens.
How does Agrobacterium-mediated gene transfer work?
Agrobacterium-mediated gene transfer works by using a modified form of Agrobacterium tumefaciens that can transfer foreign DNA into the plant cell. The foreign DNA is incorporated into the genome of the plant cell, resulting in the expression of the transferred gene.
What types of plants can be transformed using Agrobacterium-mediated gene transfer?
Agrobacterium-mediated gene transfer has been successfully used to transform a wide range of plant species, including crops, ornamental plants, and model plants.
What are the advantages of Agrobacterium-mediated gene transfer over other genetic engineering techniques?
Agrobacterium-mediated gene transfer is considered to be a highly efficient and precise method for introducing foreign genes into plants. It also allows for the transfer of larger DNA fragments compared to other methods.
What are the potential applications of Agrobacterium-mediated gene transfer?
Agrobacterium-mediated gene transfer has a wide range of potential applications, including the development of crops with improved traits such as disease resistance, increased yield, and enhanced nutritional value.
Is Agrobacterium-mediated gene transfer safe for the environment?
Agrobacterium-mediated gene transfer has been extensively studied and is considered to be safe for the environment. However, as with any genetic engineering technique, the potential risks associated with the release of genetically modified organisms into the environment must be carefully evaluated.
Can Agrobacterium-mediated gene transfer be used to transfer genes between different species?
Agrobacterium-mediated gene transfer is primarily used to transfer genes between plants of the same or closely related species. However, it is possible to use this technique to transfer genes between more distantly related species.
What are the limitations of Agrobacterium-mediated gene transfer?
Agrobacterium-mediated gene transfer is limited by the ability of the Agrobacterium to infect the target plant species. It is also limited by the availability of suitable plant tissue for transformation.
How long does it take to generate a transgenic plant using Agrobacterium-mediated gene transfer?
The time required to generate a transgenic plant using Agrobacterium-mediated gene transfer can vary depending on the target plant species and the specific gene of interest. However, it typically takes several months to generate a transgenic plant using this technique.
Are there any ethical concerns associated with Agrobacterium-mediated gene transfer?
As with any genetic engineering technique, there may be ethical concerns associated with the use of Agrobacterium-mediated gene transfer, particularly with regard to the potential impact on biodiversity and the use of genetically modified organisms in agriculture. These concerns should be carefully evaluated and addressed before using this technique in a commercial or agricultural context.
References
- Mehrotra S, Goyal V. Agrobacterium-mediated gene transfer in plants and biosafety considerations. Appl Biochem Biotechnol. 2012 Dec;168(7):1953-75. doi: 10.1007/s12010-012-9910-6. Epub 2012 Oct 23. PMID: 23090683.
- Hwang, H.-H., Yu, M., & Lai, E.-M. (2017). Agrobacterium-Mediated Plant Transformation: Biology and Applications. The Arabidopsis Book, 15, e0186. doi:10.1199/tab.0186
- Amilia Pratiwi, R., & Imam Surya, M. (2020). Agrobacterium-Mediated Transformation. IntechOpen. doi: 10.5772/intechopen.91132
- Gelvin SB. Agrobacterium-mediated plant transformation: the biology behind the “gene-jockeying” tool. Microbiol Mol Biol Rev. 2003 Mar;67(1):16-37, table of contents. doi: 10.1128/MMBR.67.1.16-37.2003. PMID: 12626681; PMCID: PMC150518.
- Tzfira, T., & Citovsky, V. (2006). Agrobacterium-mediated genetic transformation of plants: biology and biotechnology. Current Opinion in Biotechnology, 17(2), 147–154. doi:10.1016/j.copbio.2006.01.009
- Hensel, G., Kastner, C., Oleszczuk, S., Riechen, J., & Kumlehn, J. (2009). Agrobacterium-Mediated Gene Transfer to Cereal Crop Plants: Current Protocols for Barley, Wheat, Triticale, and Maize. International Journal of Plant Genomics, 2009, 1–9. doi:10.1155/2009/835608
- Hohn, B., Koukolíková-Nicola, Z., Bakkeren, G., & Grimsley, N. (1989). Agrobacterium-mediated gene transfer to monocots and dicots. Genome, 31(2), 987–993. doi:10.1139/g89-172
- https://bp.ueb.cas.cz/pdfs/bpl/2009/02/01.pdf
- https://phytopharmajournal.com/assets/pdf_files/Vol11_Issue2_11.pdf
- http://globalsciencebooks.info/Online/GSBOnline/images/0812/TPJ_2(2)/TPJ_2(2)127-137o.pdf
- https://labassociates.com/agrobacterium-mediated-plant-genetic-transformation-an-overview
- https://www.deshbandhucollege.ac.in/pdf/resources/1589512616_Z(H)-VI-Biotech-1.pdf
- https://mgcub.ac.in/pdf/material/202004260029504668c533f4.pdf
- https://goldbio.com/articles/article/a-quick-overview-of-agrobacterium-for-plant-transformation
- https://www.mybiosource.com/learn/testing-procedures/agrobacterium-mediated-gene-transfer/
- https://www.davuniversity.org/images/files/study-material/Agrobacterium-mediated%20gene%20transfer.pdf
- https://www.slideshare.net/NISHANTHSEKAR1/agrobacterium-mediated-gene-transfer-77002058
Related Posts
- Organic Waste Recycling – Definition, Characteristics, Methods, Steps, Significance
- Microbial Degradation of Lignin – Microorganisms, Enzymes, Steps, Mechanisms, Challenges
- Microbial degradation of cellulose – Enzymes, Steps, Mechanisms
- Phyllosphere Microorganisms – Examples, Factors, Effects
- Soil Formation (Pedogenesis)- Definition, Factors, Process, Steps, Examples
