Contents
What are Okazaki Fragments?
- Okazaki fragments, named after the Japanese molecular biologists Reiji and Tsuneko Okazaki who first identified them in the 1960s, are short DNA sequences synthesized during the replication of the lagging strand of DNA. These fragments are essential components in the semi-discontinuous replication process, ensuring the accurate duplication of the genetic code.
- DNA replication is a complex and highly regulated process that ensures the faithful transmission of genetic information from one generation of cells to the next. During this process, the double-stranded DNA molecule is unwound by the enzyme DNA helicase, creating a structure known as the replication fork. At this fork, the synthesis of new complementary DNA strands commences, facilitated by enzymes like DNA primase and DNA polymerase.
- However, the inherent antiparallel nature of DNA strands presents a challenge. DNA polymerases can only synthesize new DNA in the 5’ to 3’ direction. Consequently, while the leading strand is synthesized continuously in the direction of the replication fork, the lagging strand, oriented in the 5’ to 3’ direction relative to the fork, must be replicated in a discontinuous manner. This is where Okazaki fragments come into play.
- On the lagging strand, DNA primase synthesizes short RNA primers, which serve as starting points for DNA polymerase. The polymerase then extends these primers, creating Okazaki fragments. These fragments, in eukaryotic cells, typically range from 100 to 200 nucleotides in length. Once synthesized, the RNA primers are replaced with DNA, and the enzyme DNA ligase seals the gaps between the fragments, resulting in a continuous DNA strand.
- The discovery of Okazaki fragments was a significant milestone in molecular biology. Through experiments with the bacterium Escherichia coli, the Okazakis demonstrated that DNA replication on the lagging strand was not continuous, as previously believed. Instead, they identified the presence of short DNA fragments, which were later termed Okazaki fragments, as transient intermediates in the replication process.
- In summary, Okazaki fragments are pivotal in the DNA replication process, ensuring the accurate and complete replication of the lagging strand. Their discovery reshaped our understanding of DNA synthesis, highlighting the intricacies and precision of cellular processes.
Okazaki Fragments Definition
Okazaki fragments are short DNA sequences synthesized discontinuously on the lagging strand during DNA replication, which are later joined together to form a continuous strand.
Why do okazaki fragments form?
Why okazaki fragments are formed? Okazaki fragments form due to the inherent nature of DNA polymerase and the antiparallel structure of DNA. Here’s a breakdown of why Okazaki fragments form:
- Antiparallel Structure of DNA:
- DNA consists of two strands that run in opposite directions: one runs from the 5′ to 3′ direction, and the other runs from the 3′ to 5′ direction. These are often referred to as the antiparallel strands of the DNA double helix.
- Directionality of DNA Polymerase:
- DNA polymerase, the enzyme responsible for synthesizing new DNA strands, can only add nucleotides to the 3′ end of a growing DNA strand. This means that DNA polymerase can only synthesize DNA in the 5′ to 3′ direction.
- Continuous Synthesis of the Leading Strand:
- When the DNA double helix unwinds to be replicated, one of the strands (the leading strand) is oriented in such a way that its 3′ end faces the replication fork. This allows DNA polymerase to synthesize the leading strand continuously in the 5′ to 3′ direction as the fork progresses.
- Discontinuous Synthesis of the Lagging Strand:
- The other strand (the lagging strand) has its 3′ end facing away from the replication fork. Due to the 5′ to 3′ synthesis limitation of DNA polymerase, this strand cannot be replicated continuously as the leading strand. Instead, as the replication fork progresses and more of the lagging strand template is exposed, DNA synthesis on this strand must start anew from a point closer to the replication fork and proceed backward, away from the fork. This results in the formation of short, discontinuous fragments of DNA known as Okazaki fragments.
- Role of RNA Primers:
- The initiation of each Okazaki fragment synthesis begins with an RNA primer. This primer provides the initial 3′ end for DNA polymerase to start adding nucleotides. As the replication fork continues to move forward, a new RNA primer is laid down, and a new Okazaki fragment begins to form.
- Completion of the Lagging Strand:
- After the synthesis of each Okazaki fragment, the RNA primers are removed and replaced with DNA. The gaps between the fragments are then sealed by the enzyme DNA ligase, resulting in a continuous lagging strand.
In summary, Okazaki fragments form on the lagging strand during DNA replication because of the antiparallel nature of DNA and the directionality of DNA polymerase. The formation of these fragments is a clever biological solution to ensure that both strands of the DNA double helix are accurately replicated.
Discovery of Okazaki Fragments
- The discovery of Okazaki fragments marked a pivotal moment in the understanding of DNA replication. In the early 1960s, Tsuneko and Reiji Okazaki, researchers at Nagoya University in Japan, embarked on a study to elucidate the mechanisms underlying DNA replication. Their hypothesis centered on the idea that the semiconservative replication of DNA might involve a discontinuous synthesis mechanism for the daughter strands.
- The Okazakis postulated that the lagging strand, which grows in the 3’–5′ direction, might be synthesized in short segments in the 5’–3′ direction, opposite to its actual extension. By subsequently linking these segments, the daughter strand could be elongated.
- To validate this hypothesis, they employed a pulse-chase experiment. They briefly exposed actively replicating DNA to tritiated nucleotides, allowing these “hot” nucleotides to integrate into the DNA strands. Following this pulse, the DNA was rapidly isolated and exposed to “cold” unlabeled nucleotides for varying durations.
- Centrifugation of the DNA samples revealed that, after short chase durations, most of the radioactivity was associated with small DNA fragments. However, with longer chase durations, the radioactivity was found in larger DNA strands, suggesting that the small fragments were being incorporated into the larger strands over time.
- This groundbreaking experiment provided evidence for the existence of short DNA fragments during the synthesis of the lagging strand. These fragments were subsequently named “Okazaki fragments” in honor of their discoverers, a designation proposed by Rollin Hotchkiss in 1968 during the Cold Spring Harbor Symposium.
- Further supporting experiments involved pulse-labeling Escherichia coli (E.coli) with 3H-thymidine under conditions that slowed their growth and division rates. The bacteria were initially cultured at 37℃ with 14C-thymidine to uniformly label their DNA.
- Subsequently, the temperature was reduced to 20℃, and the cells were pulse-labeled with 3H-thymidine for 10 seconds. This approach aimed to identify transient intermediates in DNA replication under slowed replication conditions.
- Following the pulse, the DNA was isolated and subjected to alkaline sucrose gradient sedimentation. Analysis of the gradient fractions revealed that the majority of the 3H-labeled DNA initially appeared as fragments ranging from 50 to 5000 nucleotides in length. Over time, these fragments were elongated and integrated, serving as transient intermediates in DNA replication.
- In conclusion, the meticulous experiments conducted by the Okazakis unveiled the existence of short DNA fragments, now known as Okazaki fragments, which play a crucial role in the discontinuous synthesis of the lagging strand during DNA replication. This discovery reshaped the scientific community’s understanding of the intricate processes governing DNA replication.
Why Okazaki fragments are discontinuous?
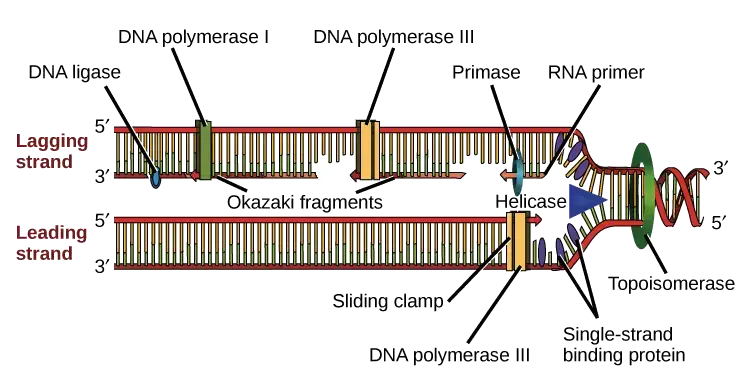
- Okazaki fragments are short, newly synthesized DNA sequences formed during the replication process. The discovery and understanding of these fragments can be attributed to the pioneering work of Kiwako Sakabe, Reiji Okazaki, and Tsuneko Okazaki. Historically, the prevailing belief in the scientific community was that DNA replication occurred continuously in both the 3′ to 5′ and 5′ to 3′ directions. The numbers 3′ and 5′ refer to specific carbons on the deoxyribose ring in nucleic acids, denoting the orientation or directionality of a DNA strand.
- In 1967, a groundbreaking proposition was made by Tsuneko Okazaki and Toru Ogawa. They posited that no known mechanism allowed for continuous replication in the 3′ to 5′ direction. Instead, replication in this direction might be discontinuous, with DNA being synthesized in short segments. These segments would then be attached in the 5′ to 3′ direction to the pre-existing strand, facilitated by the enzyme DNA polymerase.
- To validate this hypothesis, the team employed an experimental approach wherein they labeled newly replicated regions of Escherichia coli chromosomes. By denaturing and extracting the DNA, they observed a significant presence of radioactive short DNA units, suggesting a discontinuous replication mechanism. This hypothesis gained further traction with the identification of polynucleotide ligase, an enzyme responsible for connecting these short DNA strands.
- In a subsequent experiment in 1968, the Okazakis provided more compelling evidence. They postulated that if DNA synthesis indeed involved discontinuous replication, then conditions impairing the function of ligase should lead to an accumulation of these short DNA chains. Using E. coli infected with a specific bacteriophage T4, which produces a temperature-sensitive polynucleotide ligase, they observed a significant accumulation of short DNA chains at elevated temperatures. This observation not only reinforced their hypothesis but also debunked the idea that these short chains were mere artifacts of the extraction process.
- In essence, the meticulous experiments conducted by the Okazakis elucidated the discontinuous nature of DNA replication, particularly on the lagging strand. This led to the identification and understanding of what we now recognize as Okazaki fragments. These fragments play a pivotal role in ensuring accurate DNA replication, highlighting the intricacies and precision inherent in cellular processes.
Formation of Okazaki Fragments – How are okazaki fragments synthesized?
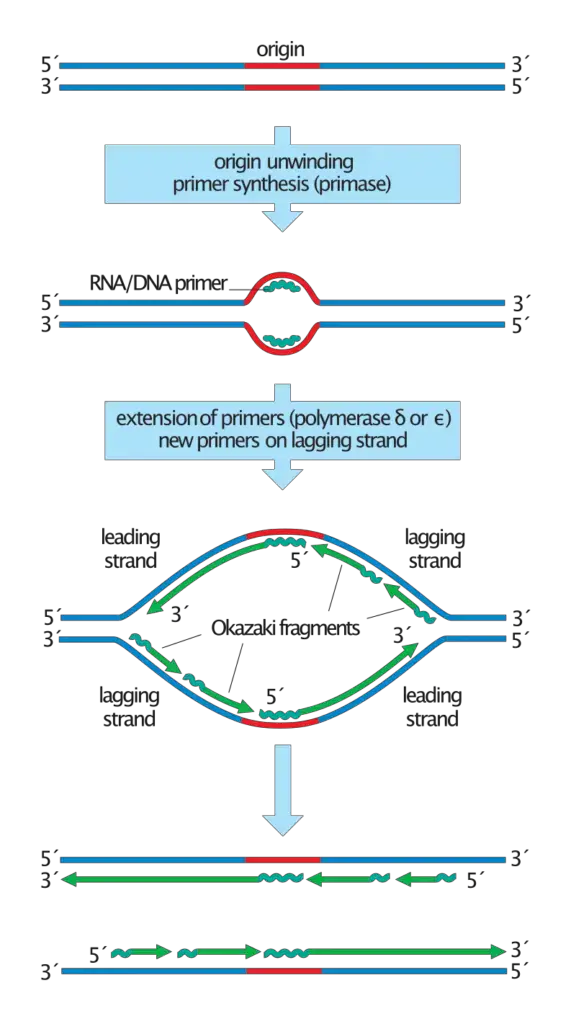
- The formation of Okazaki fragments is a fundamental aspect of DNA replication, particularly concerning the synthesis of the lagging strand. The process begins with the formation of the DNA replication fork, a structure that emerges when the DNA double helix is unwound. This unwinding is facilitated by the enzyme DNA helicase, which separates the complementary DNA strands.
- As replication progresses, enzymes like DNA primase and DNA polymerase play pivotal roles in synthesizing a new complementary strand. However, the inherent directionality of these enzymes, which can only function in the 5′ to 3′ direction, presents a challenge. For the leading strand, which has a template in the 3′ to 5′ direction, replication is continuous, with DNA polymerase synthesizing the strand in tandem with the advancing replication fork.
- Conversely, the lagging strand, with its 5′ to 3′ template directionality, cannot be synthesized continuously. Instead, its formation is characterized by intermittent breaks. The synthesis of the lagging strand is initiated by RNA primers, produced by primase.
- These primers serve as starting points for DNA polymerase δ in eukaryotes (and DNA polymerase I in prokaryotes) to extend the strand. Due to the opposing movement of the primase and polymerase relative to the replication fork, these enzymes must frequently halt and restart, a process facilitated by the DNA helicase.
- The synthesis of each Okazaki fragment necessitates a new RNA primer. The priming process unfolds in a sequential manner: firstly, the PriA protein displaces SSB proteins from a DNA segment, followed by the binding of primase (DnaG) to PriA, and finally, the synthesis of an 11-12 base RNA primer by primase.
- As the lagging strand synthesis progresses, the DNA polymerase releases its grip on the DNA, relocates to the 3′ end of a new RNA primer, and commences the synthesis of a new DNA segment. This dynamic movement is orchestrated by the clamp-loading complex, which manages the disassembly and relocation of the sliding clamp.
- Notably, only one of the two Polymerase III core enzymes in the replisome undergoes this relocation, ensuring that only the lagging strand experiences frequent clamp adjustments.
- Post-synthesis, the RNA primers must be removed to facilitate the seamless joining of Okazaki fragments. Enzymes with endonucleolytic activity, such as Ribonuclease H (RNAse H), Flap endonucleases (FENs), and Dna2 helicase/nucleases, undertake this task.
- In prokaryotes, the FEN nuclease function is embedded within DNA polymerase I, while in eukaryotes, FENs are separate entities. The exact mechanism by which RNA-DNA primers are cleared from Okazaki fragments remains an area of ongoing research.
- Once the primers are removed, DNA ligase connects the fragments using phosphodiester linkages, resulting in a continuous DNA strand. This dual-mode of replication, with one strand being synthesized continuously and the other discontinuously, is termed semi-discontinuous replication.
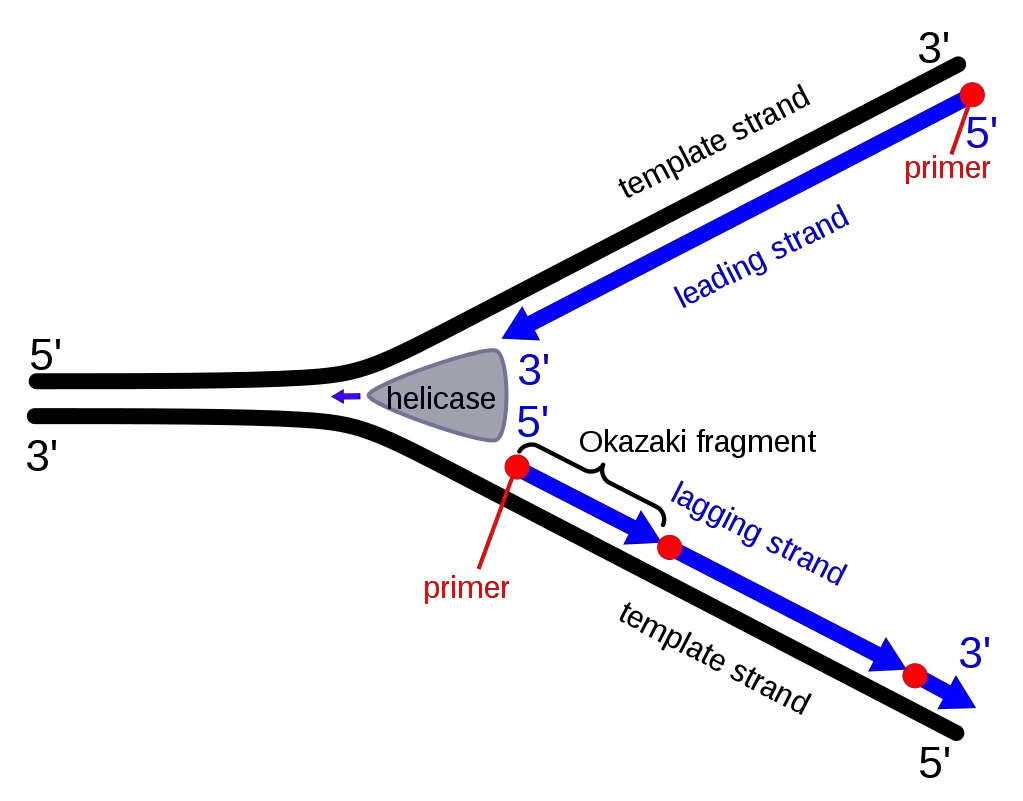
Formation of Okazaki Fragments: A Step-by-Step Process
- Initiation of the Replication Fork:
- The DNA double helix unwinds.
- The enzyme DNA helicase separates the complementary DNA strands, forming the replication fork.
- Activation of Enzymes:
- DNA primase and DNA polymerase are activated at the replication fork to begin the synthesis of a new complementary DNA strand.
- Leading Strand Synthesis:
- The leading strand, with its 3′ to 5′ template orientation, undergoes continuous replication by DNA polymerase, which follows the replication fork seamlessly.
- Lagging Strand Synthesis:
- Due to the 5′ to 3′ template orientation of the lagging strand, DNA polymerase moves in the opposite direction of the replication fork.
- This movement results in periodic interruptions in the synthesis process.
- Initiation of Okazaki Fragments with RNA Primers:
- DNA primase synthesizes an RNA primer as a starting point for each Okazaki fragment.
- DNA polymerase δ (in eukaryotes) or DNA polymerase I (in prokaryotes) extends these primers close to the replication fork.
- Synthesis of New RNA Primers:
- PriA protein displaces SSB proteins from a segment of DNA.
- Primase (DnaG) binds to PriA.
- Primase synthesizes an RNA primer consisting of 11-12 bases.
- Relocation of DNA Polymerase:
- The DNA polymerase working on the lagging strand releases its current position.
- It then relocates to the 3′ end of the new RNA primer to begin the synthesis of a new Okazaki fragment.
- Sliding Clamp Dynamics:
- The clamp-loading complex manages the disassembly and movement of the sliding clamp.
- Only one of the two Polymerase III core enzymes in the replisome releases and reattaches the sliding clamp, ensuring that only the lagging strand requires frequent clamp adjustments.
- Removal of RNA Primers:
- Enzymes with endonucleolytic activity, such as Ribonuclease H (RNAse H), Flap endonucleases (FENs), and Dna2 helicase/nucleases, remove the RNA primers.
- In prokaryotes, the FEN nuclease function is performed by a domain of DNA polymerase I.
- Ligation of Okazaki Fragments:
- Once the Okazaki fragments are synthesized and RNA primers are removed, DNA ligase joins the fragments using phosphodiester linkages.
- This results in the formation of a continuous DNA strand.
- Completion of Replication:
- Given the continuous synthesis of the leading strand and the discontinuous synthesis of the lagging strand, the overall DNA replication process is termed as semi-discontinuous.
Pathways in Okazaki Fragments Processing
The intricate process of DNA replication, particularly in the synthesis of the lagging strand, involves the formation of Okazaki fragments. The proper processing of these fragments is crucial for the continuity and integrity of the newly synthesized DNA strand. Three distinct pathways have been identified to manage the processing of these fragments:
- Short Flap Pathway: In eukaryotic DNA replication, the lagging strand is intermittently primed, leading to the formation of Okazaki fragments. The short flap pathway predominantly involves the nuclease FEN1. As DNA polymerase delta (Pol δ) synthesizes the lagging strand, it occasionally displaces the RNA/DNA initiator primer, forming a 5′ flap. FEN1, a 5’-3’ endonuclease, identifies and cleaves this flap, preparing it for subsequent ligation. This pathway ensures the removal of the Pol α-synthesized primer. The cleavage process is facilitated by the tracking mechanism of FEN1, which navigates from the 5’ flap to its base. DNA ligase then seals the nick, resulting in a continuous DNA strand. The proliferating cell nuclear antigen (PCNA) plays a pivotal role in this process, enhancing the enzymatic functions of both FEN1 and DNA ligase, ensuring the proper ligation of the lagging strand.
- Long Flap Pathway: There are instances where FEN1 temporarily disengages from the replication complex, leading to the formation of longer flaps by Pol δ. These elongated flaps, when bound by the replication protein A (RPA), necessitate the involvement of an alternate nuclease, DNA2, for processing. DNA2 collaborates with FEN1 to manage these long flaps. It can displace RPA from the flap and, using a mechanism similar to FEN1, cleave the flap, rendering it suitable for FEN1 cleavage and subsequent ligation. This pathway, termed the long flap method, highlights the versatility of the cellular machinery in managing varied replication challenges.
- Alternate Pathway: Recent research has unveiled an additional pathway for Okazaki fragment processing. This alternate mechanism involves the combined action of Pol δ and Pif1, which collaboratively execute the flap removal process, akin to the combined action of Pol δ and FEN1. This discovery underscores the complexity and adaptability of the DNA replication process, ensuring the fidelity and continuity of genetic information across generations.
Okazaki Fragments Function
Okazaki fragments play a pivotal role in the intricate process of DNA replication, ensuring the accurate and complete duplication of genetic information, which is fundamental for cell growth and division.
- Facilitation of Lagging Strand Synthesis:
- DNA replication is a bidirectional process, with one strand (the leading strand) being synthesized continuously and the other (the lagging strand) being synthesized discontinuously. Okazaki fragments are integral to the synthesis of the lagging strand. They allow DNA polymerase to synthesize the lagging strand in the 5′ to 3′ direction, even though its natural orientation is opposite to the direction of replication.
- Replacement of RNA Primers with DNA:
- The initiation of each Okazaki fragment begins with an RNA primer. DNA polymerase I, a specific type of DNA polymerase, is responsible for removing these RNA primers and replacing them with DNA. This ensures that the final replicated DNA strand is composed entirely of DNA nucleotides.
- Formation of Continuous DNA Strands:
- Once the Okazaki fragments are synthesized and the RNA primers are replaced with DNA, it is crucial to join these fragments to form a continuous DNA strand. DNA ligase accomplishes this by sealing the gaps in the sugar-phosphate backbone of the fragments, resulting in two continuous and identical daughter DNA strands.
- Maintenance of Genomic Integrity:
- Proper processing of Okazaki fragments is paramount for maintaining the integrity of the genome. Defects in the formation or processing of these fragments can lead to DNA strand breaks and chromosomal abnormalities. Such anomalies can manifest as changes in chromosome appearance, alterations in chromosome numbers, or even changes in the number of chromosome sets.
- Support in Cell Division and Growth:
- DNA replication, facilitated by the proper formation and processing of Okazaki fragments, is a prerequisite for cell division. In unicellular organisms, this division serves as a mode of asexual reproduction. In multicellular organisms, cell division is essential for growth, repair, and the generation of cells necessary for sexual reproduction. Ensuring that both daughter cells receive identical genetic material is crucial for the continuity of life and the preservation of genetic information.
Enzymes involved in Okazaki fragments formation
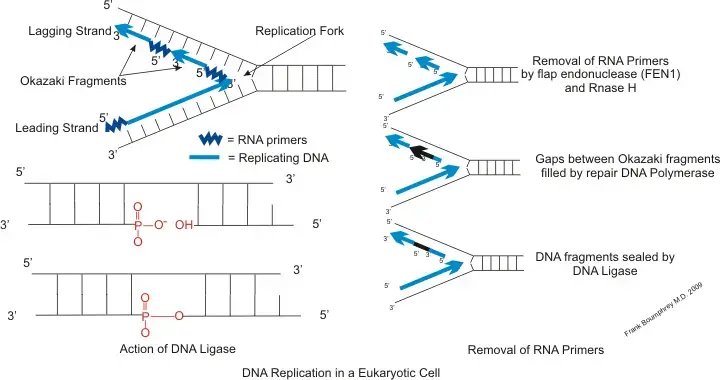
- Primase:
- Primase is responsible for adding RNA primers to the lagging strand, facilitating the synthesis of Okazaki fragments from 5′ to 3′. Due to the slower rate of RNA primer creation by primase compared to DNA polymerase’s synthesis rate, primase acts as a temporary stop signal. This halts the replication fork’s progression momentarily, preventing the leading strand from surpassing the lagging strand.
- DNA Polymerase δ:
- DNA polymerase plays a crucial role in synthesizing both the leading and lagging strands. The synthesis involves three phases with two distinct polymerases: DNA polymerase α-primase and DNA polymerase δ. The process commences with the displacement of the RNA and DNA primer by the clamp loader replication effect, leading to the attachment of the sliding clamp onto the DNA. Subsequently, DNA polymerase δ synthesizes until it reaches the 5’ end of the preceding Okazaki fragment. This enzyme also supplements the FEN1/RAD27 5’ Flap Endonuclease activity, ensuring the proper processing of Okazaki fragments.
- DNA Ligase I:
- DNA ligase I is pivotal in connecting the Okazaki fragments during lagging strand synthesis, especially after the RNA primers are replaced with DNA by DNA polymerase δ. Proper ligation is essential to prevent potential double-strand breaks in the DNA. Additionally, proliferating cell nuclear antigen (PCNA) plays a supplementary role, aiding DNA ligase I in joining the fragments.
- Flap Endonuclease 1 (FEN1):
- FEN1 is tasked with processing the Okazaki fragments. It collaborates with DNA polymerase to eliminate the RNA primer of an Okazaki fragment and can also remove the 5′ ribonucleotide and 5′ flaps during lagging strand synthesis. This process, termed nick translation, prepares the fragments for ligation.
- Dna2 Endonuclease:
- Dna2 endonuclease, although not structurally specific, is vital for cleaving long DNA flaps left by FEN1 during the Okazaki fragment maturation process. It is essential for removing the initiator RNA segment on the fragments. Additionally, Dna2 plays a role in various DNA metabolisms and is involved in telomere maintenance. Its activity is particularly significant when RNA segments attach at the 5’ end, as it operates in the 5’ to 3’ direction. In the presence of the single-stranded DNA-binding protein RPA, when DNA 5′ flaps become extended, Dna2 reduces the 3′ end of these fragments, enabling FEN1 to efficiently process the flaps.
In summary, the formation of Okazaki fragments during DNA replication is a complex process involving multiple enzymes. Each enzyme plays a specific and crucial role in ensuring the accurate and efficient synthesis of the lagging strand, safeguarding the integrity of the genome.
What enzyme joins okazaki fragments?
The enzyme that joins Okazaki fragments is DNA ligase. It seals the gaps between the Okazaki fragments, creating a continuous DNA strand on the lagging strand during DNA replication.
Mechanism of DNA Ligase Joining Okazaki Fragments:
- Recognition of the Nick:
- During DNA replication, the lagging strand is synthesized discontinuously as Okazaki fragments. Each fragment starts with an RNA primer, which is later replaced by DNA. This leaves a nick, which is a site where there’s a break in the sugar-phosphate backbone of the DNA, specifically between the 3′-OH group of one nucleotide and the 5′-phosphate group of the next nucleotide.
- Activation of DNA Ligase:
- DNA ligase first becomes activated by ATP (in eukaryotes) or NAD+ (in some prokaryotes). This results in the formation of a covalent bond between a lysine residue in the ligase and the AMP portion of ATP, releasing pyrophosphate. The ligase is now “charged” and ready to act.
- Formation of a Phosphodiester Bond:
- The activated ligase enzyme then transfers the AMP to the 5′-phosphate group at the nick site. This creates a reactive intermediate where the DNA is adenylated.
- The 3′-OH group of the nucleotide adjacent to the nick then attacks the adenylated phosphate group. This results in the formation of a phosphodiester bond, effectively sealing the nick.
- AMP is released in the process.
- Completion:
- The nick in the DNA backbone is now sealed, and the DNA ligase resets, ready to act on the next available nick.
This action of DNA ligase ensures that the lagging strand, after being synthesized as a series of Okazaki fragments, becomes a continuous DNA strand, matching the continuous synthesis observed on the leading strand.
Differences of prokaryotes and eukaryotes Okazaki fragments
- Origins of Replication:
- Prokaryotic DNA typically has a single origin of replication, whereas eukaryotic DNA molecules are larger and usually have multiple origins of replication. This results in each eukaryotic chromosome being composed of numerous replicating units of DNA, in contrast to the singular replicating unit in prokaryotes.
- Replication Environment:
- In prokaryotes, replication occurs within the cytoplasm. In contrast, eukaryotic DNA replication takes place inside the nucleus.
- Clamp Loader Complex and Clamps:
- Eukaryotes possess a clamp loader complex and a six-unit clamp known as the proliferating cell nuclear antigen. This efficient movement of the replication fork ensures that the generation of Okazaki fragments can keep pace with the continuous synthesis of DNA on the leading strand. Such clamp loader complexes are characteristic of all eukaryotes and highlight some of the nuanced differences between prokaryotic and eukaryotic Okazaki fragment synthesis.
- Length of Okazaki Fragments:
- Prokaryotic Okazaki fragments, especially in E. coli, can be as long as 2,000 nucleotides. In contrast, eukaryotic Okazaki fragments are typically shorter, ranging from 100 to 200 nucleotides.
- Replication Speed:
- Replication in prokaryotes is notably faster than in eukaryotes. For instance, bacterial replication can sometimes complete in just 40 minutes, while eukaryotic animal cells might take up to 400 hours.
- Chromosomal Structure:
- Prokaryotes have circular chromosomes, which means they lack chromosome ends to replicate. Eukaryotes, on the other hand, have linear chromosomes and have developed a unique mechanism to replicate the telomeres at their ends.
- Replication Timing:
- Prokaryotic replication is a continuous process, whereas eukaryotic cells only undergo DNA replication during the S-phase of the cell cycle.
- DNA Quantity:
- Eukaryotic cells typically contain about 25 times more DNA than prokaryotic cells.
In summary, while both prokaryotes and eukaryotes utilize Okazaki fragments in the DNA replication process, the mechanisms, structures, and intricacies of their formation and processing differ significantly between the two domains of life. These differences underscore the evolutionary complexities and adaptations that have arisen in response to the distinct cellular architectures and life cycles of prokaryotes and eukaryotes.
| Feature | Prokaryotes | Eukaryotes |
|---|---|---|
| Origins of Replication | Single origin of replication. | Multiple origins of replication, leading to numerous replicating units of DNA in each chromosome. |
| Replication Environment | Occurs within the cytoplasm. | Takes place inside the nucleus. |
| Clamp Loader Complex | Absent. | Present, with a six-unit clamp known as the proliferating cell nuclear antigen. |
| Length of Okazaki Fragments | Longer fragments, up to 2,000 nucleotides in E. coli. | Shorter fragments, typically ranging from 100 to 200 nucleotides. |
| Replication Speed | Faster, can complete in about 40 minutes. | Slower, can take up to 400 hours in animal cells. |
| Chromosomal Structure | Circular chromosomes, lacking chromosome ends to replicate. | Linear chromosomes with unique mechanisms to replicate the telomeres at their ends. |
| Replication Timing | Continuous replication process. | DNA replication occurs only during the S-phase of the cell cycle. |
| DNA Quantity | Contains significantly less DNA, typically 25 times less than eukaryotic cells. | Contains about 25 times more DNA than prokaryotic cells. |
MCQ Quiz
What are Okazaki fragments primarily associated with?
a) Transcription of RNA
b) Replication of the leading strand of DNA
c) Replication of the lagging strand of DNA
d) Translation of proteins
Which enzyme is responsible for synthesizing the RNA primers that initiate Okazaki fragments?
a) DNA polymerase
b) DNA ligase
c) Primase
d) Helicase
Which enzyme is responsible for joining Okazaki fragments together?
a) DNA polymerase
b) DNA ligase
c) Primase
d) Helicase
On which strand of the DNA do Okazaki fragments form?
a) Leading strand
b) Lagging strand
c) Both strands
d) Neither strand
Why do Okazaki fragments form during DNA replication?
a) Because DNA is single-stranded
b) Due to the antiparallel nature of DNA and the directionality of DNA polymerase
c) To repair damaged DNA
d) To assist in DNA transcription
Which of the following enzymes is NOT directly involved in the processing of Okazaki fragments?
a) DNA ligase
b) Primase
c) RNA polymerase
d) DNA polymerase
Approximately how long are Okazaki fragments in eukaryotes?
a) 10-50 nucleotides
b) 100-200 nucleotides
c) 500-1000 nucleotides
d) 2000-5000 nucleotides
Which enzyme unwinds the DNA double helix, creating a need for Okazaki fragments on the lagging strand?
a) DNA ligase
b) DNA gyrase
c) DNA helicase
d) DNA topoisomerase
After the RNA primers of Okazaki fragments are removed, they are replaced by:
a) Proteins
b) Lipids
c) DNA
d) RNA
Which of the following best describes the synthesis of the lagging strand during DNA replication?
a) Continuous and smooth
b) Discontinuous and in fragments
c) In a 3′ to 5′ direction
d) Without the need for primers
FAQ
What are the Okazaki fragments?
Okazaki fragments are short sequences of DNA nucleotides synthesized discontinuously on the lagging strand during DNA replication.
What is the function of the Okazaki fragments?
The function of Okazaki fragments is to enable the synthesis of the lagging strand of DNA in the 5′ to 3′ direction, consistent with the directionality of DNA polymerase.
What are Okazaki fragments and why are they formed?
Okazaki fragments are short DNA sequences formed on the lagging strand during DNA replication. They are formed due to the antiparallel nature of DNA and the directionality of DNA polymerase, which can only synthesize DNA in the 5′ to 3′ direction.
Why Okazaki fragments are formed?
They are formed to facilitate the synthesis of the lagging strand in the 5′ to 3′ direction during DNA replication.
Where are Okazaki fragments found?
Okazaki fragments are found on the lagging strand of the DNA during replication.
Why Okazaki fragments are discontinuous?
Okazaki fragments are discontinuous because the lagging strand is synthesized in short segments, opposite to the direction of the replication fork.
Which Okazaki fragment was made first?
The Okazaki fragment closest to the replication origin was made first.
What are the Okazaki fragments in DNA chain growth?
In DNA chain growth, Okazaki fragments represent the short, discontinuous segments of DNA synthesized on the lagging strand.
Are Okazaki fragments important?
Yes, Okazaki fragments are crucial for the accurate replication of the lagging strand of DNA.
How many Okazaki fragments are there?
The number of Okazaki fragments varies depending on the length of the lagging strand being replicated. There can be hundreds to thousands of Okazaki fragments formed during the replication of a single DNA molecule.
Is an Okazaki fragment DNA or RNA?
An Okazaki fragment is DNA, but it initially starts with an RNA primer which is later replaced by DNA.
References
- Reha-Krantz, L. J. (2013). Okazaki Fragment. Brenner’s Encyclopedia of Genetics, 158–160. doi:10.1016/b978-0-12-374984-0.01087-1
- Pelley, J. W. (2012). Organization, Synthesis, and Repair of DNA. Elsevier’s Integrated Review Biochemistry, 125–135. doi:10.1016/b978-0-323-07446-9.00015-5
- Balakrishnan L, Bambara RA. Okazaki fragment metabolism. Cold Spring Harb Perspect Biol. 2013 Feb 1;5(2):a010173. doi: 10.1101/cshperspect.a010173. PMID: 23378587; PMCID: PMC3552508.
- https://www.chemeurope.com/en/encyclopedia/Okazaki_fragment.html
- https://www.rcsb.org/structure/1ofx
- https://thebumblingbiochemist.com/uncategorized/okazaki-fragments-dna-replication/
- https://www.shaalaa.com/question-bank-solutions/what-are-okazaki-fragments-dna-replication_226369
- https://www.biologyonline.com/dictionary/okazaki-fragment
- https://ib.bioninja.com.au/higher-level/topic-7-nucleic-acids/71-dna-structure-and-replic/okazaki-fragments.html
- http://www.biologyaspoetry.com/terms/okazaki_fragments.html
- https://en.wikipedia.org/wiki/Okazaki_fragments
- https://www.biorxiv.org/content/10.1101/384503v1
Related Posts
- DNA Library – Types, Construction, Applications
- Emulsion PCR – Principle, Procedure, Advantages, Limitations, Uses
- TaqMan Probe – Definition, Principle, Applications
- Differences Between Sensitivity, Specificity, False positive, False negative
- Protein Synthesis (Translation)- Definition, Steps, Sites, Machinery