Contents
What is Next-Generation Sequencing (NGS)?
Next-Generation Sequencing (NGS), also known as high-throughput sequencing, has revolutionized the field of genomics and molecular biology by allowing the sequencing of thousands to millions of DNA molecules simultaneously. It encompasses a range of different sequencing technologies, all aimed at producing large amounts of sequence data at a lower cost compared to traditional methods.
The development of NGS was driven by the high demand for affordable sequencing technologies. The goal was to overcome the limitations of conventional DNA sequencing methods, such as Sanger sequencing, which were expensive and time-consuming. NGS technologies have successfully achieved this objective, enabling faster and more cost-effective sequencing of DNA and RNA.
NGS is categorized into different generations based on the sequencing technologies used:
- First Generation: This includes the widely used Sanger sequencing, which was the first method capable of sequencing DNA. Although it provided significant insights into genomics, it was limited in terms of scalability and cost-effectiveness.
- Second Generation Sequencing: This generation encompasses several technologies, such as pyrosequencing, sequencing by reversible terminator chemistry, and sequencing by ligation. These methods allowed for higher throughput sequencing, generating thousands to millions of sequences simultaneously. They marked a significant leap forward in terms of speed and cost reduction.
- Third Generation Sequencing: The third generation of NGS introduced technologies like single molecule fluorescent sequencing, single molecule real-time sequencing, semiconductor sequencing, and nanopore sequencing. These methods offered the ability to sequence DNA molecules directly and in real-time, without the need for amplification or labeling. They provided advantages in terms of long-read sequencing, detecting modifications in DNA, and facilitating the study of complex genomic regions.
- Fourth Generation Sequencing: The concept of fourth-generation sequencing aims to conduct genomic analysis directly within the cell. This represents an emerging area of research where new technologies are being developed to enable in situ sequencing, providing insights into cellular genomics and spatial organization.
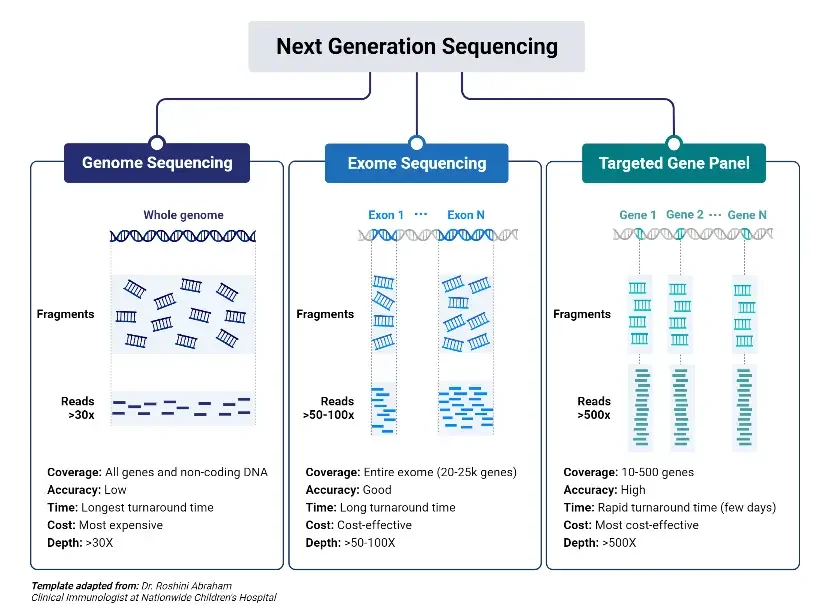
The wide range of NGS technologies and their continuous evolution have expanded the applications of sequencing in molecular biology. NGS has found usage in various fields, including genome sequencing, transcriptomics, epigenetics, metagenomics, and personalized medicine. It has enabled researchers to study complex genetic variations, identify disease-causing mutations, investigate gene expression profiles, and explore microbial diversity, among many other applications.
In summary, Next-Generation Sequencing has transformed the field of genomics and molecular biology by enabling fast and cost-effective sequencing of DNA and RNA. Its different generations of sequencing technologies have overcome the limitations of traditional methods and opened up new avenues for research, leading to breakthroughs in understanding genetics, diseases, and biological systems.
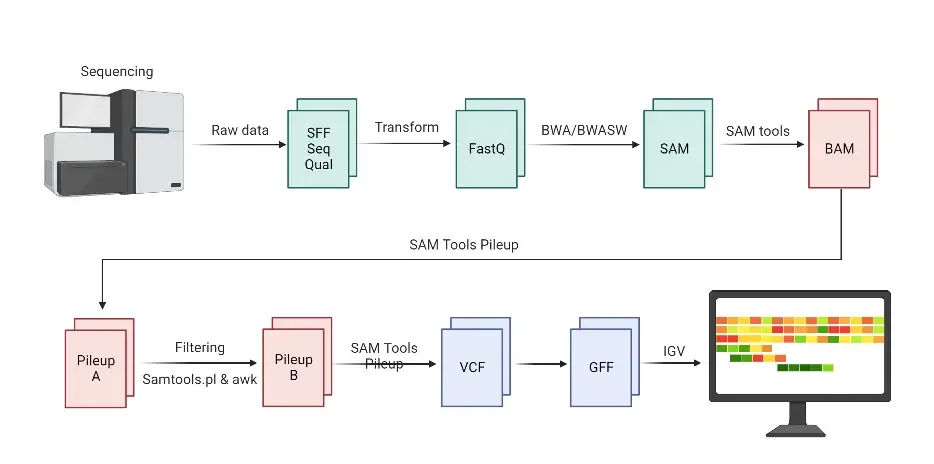
Next-Generation Sequencing workflow
The Next-Generation Sequencing (NGS) workflow involves several key steps, from constructing the sequencing library to analyzing the generated data. Here is an overview of the NGS workflow based on the provided information:
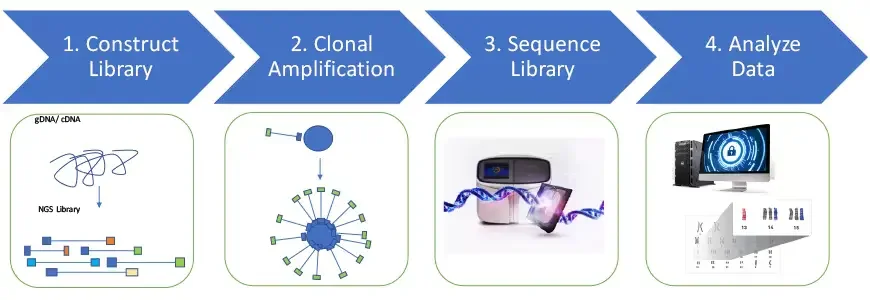
1. Construct Library
The process begins with creating a sequencing library from the DNA or cDNA sample. The DNA sample is fragmented into relatively short double-stranded fragments using methods like physical shearing, enzyme digestion, or PCR amplification of specific genetic regions. Adaptor sequences specific to the sequencing technology are then ligated to the DNA fragments, forming a fragment library. Adaptors may also include unique molecular barcodes to tag each sample, enabling pooling or multiplexing of multiple samples for simultaneous sequencing.
Additionally, there are specialized library preparation methods like paired-end libraries and mate-pair libraries. Paired-end libraries allow sequencing of DNA fragments from both ends, enhancing mapping and enabling the detection of genomic rearrangements and RNA gene fusions. Mate-pair libraries involve larger-sized DNA inserts and generate reads that are distal to each other and in opposite orientations, facilitating de novo assembly and identification of complex genomic rearrangements.
2. Clonal Amplification
Once the library is constructed, the DNA fragments need to be attached to a solid surface and clonally amplified to increase the signal for detection during sequencing. This process involves binding each unique DNA molecule in the library to beads or a flow-cell and PCR amplifying them to create a set of identical clones. This step ensures sufficient copies of the DNA fragments for sequencing.
3. Sequence Library
The amplified DNA library is then subjected to sequencing using a sequencing instrument. Different NGS technologies employ variations of the “sequencing by synthesis” method, where individual bases are read as they are incorporated into a growing polymerized strand. This process involves cycles of DNA base synthesis, detection of incorporated bases, and removal of reactants to initiate the next cycle.
Sequencing instruments typically use optical or electrical detection methods. Optical detection is commonly used and involves the detection of fluorescent signals to determine nucleotide incorporation. Electrical detection, as used in Ion Torrent technology, senses the release of hydrogen ions during nucleotide incorporation. These sequencing cycles generate large amounts of sequencing data.
4. Analyze Data
The analysis of NGS data is a crucial step in the workflow and involves several stages. Primary analysis processes the raw signals from the instrument detectors into digitized data or base calls. This step results in files containing base calls assembled into sequencing reads (FASTQ files) and their associated quality scores (Phred quality score).
Secondary analysis includes read filtering and trimming based on quality, alignment of reads to a reference genome or assembly of reads for novel genomes, and variant calling. The output of secondary analysis is a BAM file containing aligned reads. Tertiary analysis, the most challenging step, involves interpreting results and extracting meaningful information from the data.
NGS data analysis is typically performed by bioinformatics specialists due to its complexity and associated algorithms. However, user-friendly software has been developed to simplify the analysis and enable users without specialized bioinformatics training to obtain results.
In conclusion, the NGS workflow involves constructing the sequencing library, clonal amplification of DNA fragments, sequencing the library, and analyzing the generated data through primary, secondary, and tertiary analysis steps. Each step contributes to the successful acquisition and interpretation of NGS data for various applications in genomics and molecular biology.
Types of Next Generation Sequencing
Next-Generation Sequencing (NGS) has witnessed the development of various sequencing technologies that have greatly advanced the field of genomics and molecular biology. Here are some notable NGS technologies:
- Lynx Therapeutics’ Massively Parallel Signature Sequencing (MPSS): MPSS was a sequencing technology developed by Lynx Therapeutics, now part of Illumina. It involved the creation of millions of DNA fragments that were individually attached to microbeads and sequenced in parallel using DNA polymerase. MPSS provided highly accurate sequence data and was particularly useful for gene expression analysis and profiling.
- Polony Sequencing: Polony sequencing, pioneered by George Church’s lab, involved amplifying DNA fragments on a solid surface, such as a glass slide. By attaching DNA fragments to spatially separate spots, each spot representing a “polony,” multiple sequences could be determined simultaneously. Polony sequencing allowed for high-throughput sequencing and was used for applications like resequencing and whole-genome sequencing.
- Pyrosequencing: Pyrosequencing is a widely used NGS technology developed by 454 Life Sciences (Roche). It involves the detection of pyrophosphate released during DNA synthesis. By monitoring the release of pyrophosphate, the sequence of the DNA template can be determined. Pyrosequencing offered longer read lengths and was valuable for applications like de novo sequencing, targeted resequencing, and SNP detection.
- Illumina (Solexa) Sequencing: Illumina sequencing, originally developed by Solexa, later acquired by Illumina, is one of the most popular NGS technologies. It utilizes reversible dye-terminator chemistry to sequence millions of DNA fragments in parallel. Illumina sequencing platforms, such as the HiSeq and NovaSeq systems, are highly scalable, offer high throughput, and provide short read lengths. They are widely used for diverse applications, including whole-genome sequencing, exome sequencing, transcriptomics, and epigenomics.
- SOLiD Sequencing: SOLiD (Sequencing by Oligonucleotide Ligation and Detection) sequencing, developed by Applied Biosystems (Life Technologies, now part of Thermo Fisher Scientific), utilizes a unique ligation-based sequencing approach. It involves the ligation of fluorescently labeled oligonucleotides to DNA fragments, followed by multiple rounds of sequencing by ligation. SOLiD sequencing offered high accuracy and was employed for applications like whole-genome sequencing, resequencing, and DNA methylation analysis.
- DNA Nanoball Sequencing: DNA nanoball sequencing, developed by Complete Genomics (later acquired by BGI), involved the amplification of DNA fragments in a dense array of nanoballs. Each nanoball contained multiple copies of the same DNA fragment, which were subsequently sequenced in parallel. This technology offered long read lengths, high accuracy, and was used for whole-genome sequencing.
- Heliscope Single Molecule Sequencing: Heliscope sequencing, developed by Helicos Biosciences, was a single-molecule sequencing technology. It involved the immobilization of single DNA molecules on a glass surface, followed by iterative sequencing-by-synthesis. Heliscope sequencing offered the advantage of single-molecule detection and was useful for applications like whole-genome sequencing, transcriptomics, and targeted resequencing.
- Single Molecule SMRT Sequencing: Single Molecule, Real-Time (SMRT) sequencing is a technology developed by Pacific Biosciences (PacBio). It utilizes zero-mode waveguides (ZMWs) that confine DNA polymerase to observe the incorporation of fluorescently labeled nucleotides in real-time. SMRT sequencing provides long read lengths and is particularly advantageous for applications requiring the detection of structural variations, epigenetic modifications, and long-range genomic information.
- Single Molecule Real Time (RNAP) Sequencing: Single Molecule Real-Time (RNAP) sequencing, also developed by Pacific Biosciences, is a variation of SMRT sequencing. Instead of DNA polymerase, it employs RNA polymerase (RNAP) to read DNA templates, allowing for the direct sequencing of RNA molecules. RNAP sequencing is used for applications like transcriptomics, isoform discovery, and studying RNA dynamics.
These diverse NGS technologies have significantly expanded our understanding of genomics, molecular biology, and their applications in fields like personalized medicine, agriculture, evolutionary biology, and more.
Lynx therapeutics’ massively parallel signature sequencing (MPSS)
- Lynx Therapeutics’ Massively Parallel Signature Sequencing (MPSS) technology was one of the pioneering “next-generation” sequencing technologies. Developed in the 1990s at Lynx Therapeutics, a company founded by Sydney Brenner and Sam Eletr in 1992, MPSS revolutionized the field of high-throughput sequencing.
- MPSS was designed as an ultra-high-throughput sequencing technology that provided valuable insights into gene expression profiles. When applied to expression profiling, MPSS enabled the identification and quantification of nearly every transcript present in a sample, accurately measuring its expression level.
- The MPSS method was bead-based and utilized a sophisticated approach involving adapter ligation and decoding. The sequence was read in increments of four nucleotides, which allowed for the detection of hundreds of thousands of short DNA sequences. These sequences were often used for sequencing cDNA and assessing gene expression levels.
- One of the challenges with MPSS was its susceptibility to sequence-specific bias or loss of specific sequences. This meant that certain sequences could be underrepresented or missed altogether due to the nature of the method. Despite this limitation, the essential properties of the MPSS output were characteristic of later “next-gen” data types, paving the way for the development of subsequent sequencing technologies.
- MPSS played a crucial role in advancing genomic research, particularly in the study of gene expression. Its ability to generate high-throughput data and accurately quantify gene expression levels provided researchers with valuable information for understanding biological processes and diseases.
- Although MPSS is no longer widely used today, its impact on the field of next-generation sequencing cannot be overstated. It laid the foundation for the development of newer and more advanced sequencing technologies, driving progress in genomics and molecular biology.
Polony sequencing
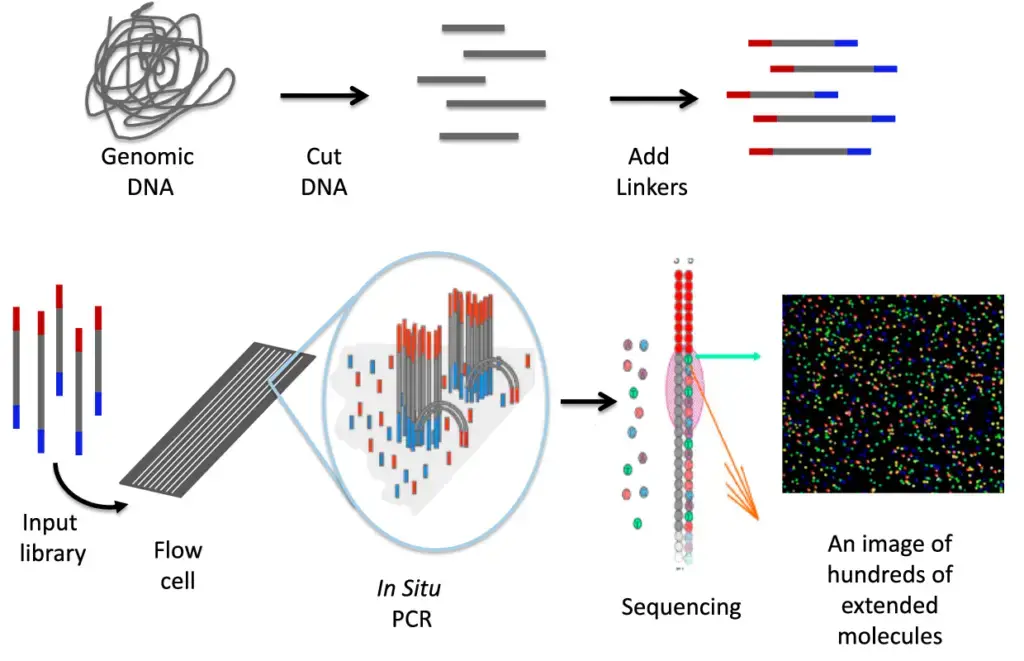
- Polony sequencing is an innovative and cost-effective multiplex sequencing technique that enables the parallel reading of millions of immobilized DNA sequences with high accuracy. The development of this technique can be attributed to Dr. George Church at Harvard Medical College.
- Polony sequencing involves the integration of several components to achieve its sequencing capabilities. It utilizes an in vitro paired-tag library, emulsion PCR, an automated microscope, and ligation-based sequencing chemistry. By combining these elements, polony sequencing enables the accurate sequencing of DNA.
- One of the notable achievements of polony sequencing was the successful sequencing of an E. coli genome with exceptional accuracy. The technique achieved a sequencing accuracy exceeding 99.9999%. This high level of accuracy was coupled with a significant cost reduction, making polony sequencing approximately one-tenth the cost of Sanger sequencing.
- The key principle behind polony sequencing is the immobilization of DNA sequences. Through emulsion PCR, individual DNA fragments are amplified within water-in-oil droplets, resulting in the formation of discrete DNA colonies or “polonies.” Each polony contains clonally amplified DNA fragments with unique barcodes or tags.
- To sequence these immobilized DNA sequences, ligation-based chemistry is employed. The sequencing chemistry involves iterative rounds of ligation, fluorescent labeling, imaging, and cleavage. The fluorescently labeled nucleotides are sequentially added to the polonies, and the incorporated nucleotides are detected using an automated microscope. This process allows for the reading of the DNA sequences within each polony in parallel.
- Polony sequencing offers several advantages, including its cost-effectiveness and high accuracy. The ability to read millions of DNA sequences simultaneously greatly enhances the throughput of the sequencing process. This makes it a valuable tool in various applications, such as genome sequencing, genetic research, and diagnostics.
- While polony sequencing has contributed to the field of genomics, it has been largely superseded by newer sequencing technologies. However, its development and success paved the way for the advancement of multiplex sequencing techniques, inspiring further innovations in the field.
SOLiD sequencing
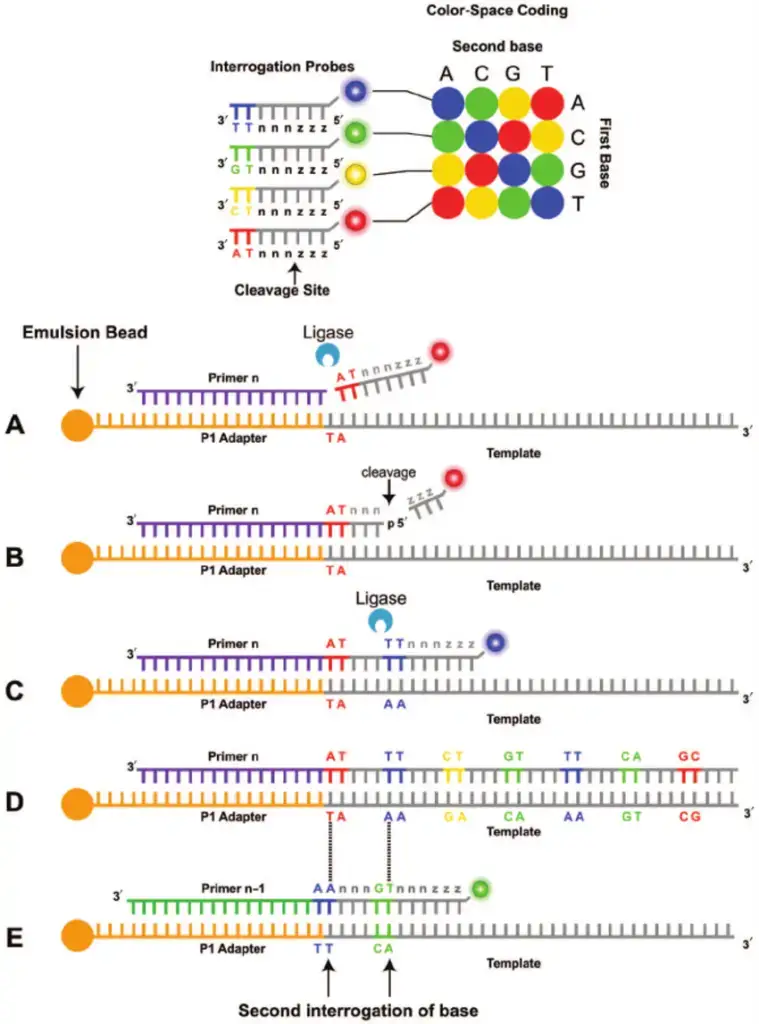
- SOLiD sequencing, developed by Applied Biosystems (now part of Thermo Fisher Scientific), is a sequencing technology that utilizes oligonucleotide ligation and detection. It offers an alternative approach to DNA sequencing and has its own unique characteristics.
- In SOLiD sequencing, a pool of oligonucleotides of fixed length is generated, representing all possible sequences at a specific position in the DNA molecule. These oligonucleotides are labeled according to the nucleotide they correspond to in the sequenced position.
- The sequencing process involves the ligation of these labeled oligonucleotides to the DNA template. Each oligonucleotide is ligated one at a time, allowing for the determination of the nucleotide at each position in the DNA sequence. The ligation events are detected and recorded, providing information about the DNA sequence.
- SOLiD sequencing generates sequencing data with quantities and lengths comparable to Illumina sequencing, another widely used sequencing technology. It offers high-throughput sequencing capabilities, allowing for the analysis of large numbers of DNA molecules simultaneously.
- The data generated from SOLiD sequencing can be used for a variety of applications, including genome sequencing, genetic variation analysis, and gene expression profiling. It has contributed to advancements in fields such as genomics, personalized medicine, and cancer research.
- While SOLiD sequencing provided valuable insights into DNA sequencing and analysis, it has been largely replaced by newer sequencing technologies, such as Illumina’s sequencing by synthesis and Pacific Biosciences’ single-molecule real-time (SMRT) sequencing. These newer technologies offer improved read lengths, scalability, and cost-effectiveness.
- Despite its reduced usage in recent years, SOLiD sequencing played a significant role in advancing the field of DNA sequencing and contributed to our understanding of genetic variation and gene expression. Its development and utilization helped pave the way for the continued progress in high-throughput sequencing technologies.
Helioscope single molecule sequencing
- Helioscope single molecule sequencing is a sequencing technology that utilizes DNA fragments with added polyA tail adapters. These DNA fragments are attached to the surface of a flow cell, which serves as the platform for sequencing.
- The sequencing process in Helioscope involves extension-based sequencing with cyclic washes of the flow cell using fluorescently labeled nucleotides. During each cycle, a labeled nucleotide is incorporated into the growing DNA strand, and the fluorescence signal is detected and recorded.
- The reads, or sequences of the DNA fragments, are generated by the Helioscope sequencer. This sequencing platform is designed to handle the specific requirements of Helioscope single molecule sequencing, ensuring accurate and reliable data generation.
- One characteristic of Helioscope sequencing is that the reads are relatively short, typically up to 55 bases per run. However, recent improvements in the methodology have allowed for more accurate reads of homopolymers, which are stretches of the same nucleotide in the DNA sequence. Additionally, Helioscope sequencing has been adapted for RNA sequencing, enabling the study of gene expression and transcriptomics.
- The short read length in Helioscope sequencing can present challenges when it comes to genome assembly and analysis, as longer reads are generally preferred for comprehensive genomic investigations. Nonetheless, Helioscope sequencing offers advantages in terms of accuracy and the ability to capture specific genomic features, such as homopolymers and RNA transcripts.
- Helioscope single molecule sequencing has found applications in various research areas, including genomics, transcriptomics, and targeted sequencing. Its ability to generate accurate reads and provide insights into specific genomic regions or RNA expression profiles makes it a valuable tool in molecular biology research.
- Continued advancements and refinements in Helioscope sequencing technology are expected to further enhance its capabilities and expand its applications in the future.
Illumina (Solexa) sequencing
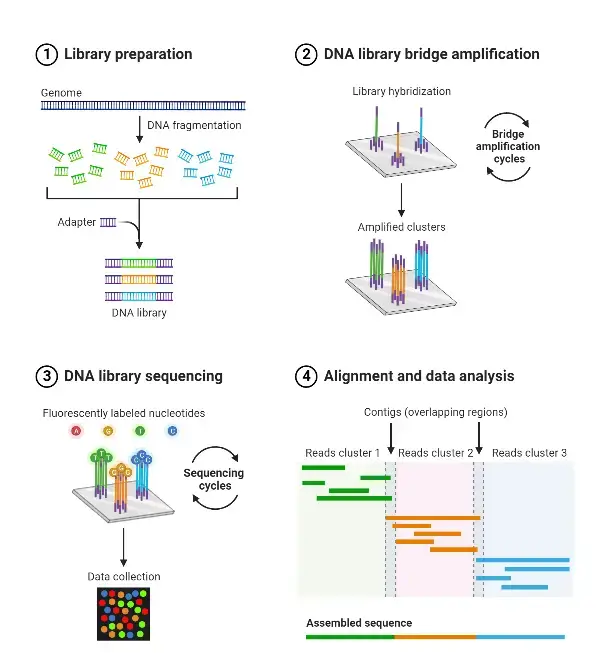
- Illumina (formerly Solexa) sequencing is a highly popular next-generation sequencing technology that is widely used for DNA sequencing applications. This sequencing method is based on the use of dye terminators to determine the nucleotide sequence of DNA molecules.
- In Illumina sequencing, the DNA molecules are first attached to primers on a slide and undergo a process called bridge amplification. During bridge amplification, the DNA molecules are amplified to create clusters, where multiple copies of the same DNA fragment are localized in a small area on the slide.
- Unlike some other sequencing methods like pyrosequencing, Illumina sequencing extends the DNA sequence by adding one nucleotide at a time. Each nucleotide is labeled with a fluorescent dye, and a camera takes images of the nucleotides during each sequencing cycle.
- After the imaging step, the fluorescent dye, along with the terminal 3′ blocker, is chemically removed from the DNA. This allows for the next sequencing cycle to commence, where a new nucleotide is added and imaged. This iterative process is repeated multiple times to obtain the complete DNA sequence.
- The images obtained from the fluorescently labeled nucleotides are analyzed to determine the sequence of the DNA fragments. The intensity of the fluorescence signal at each position indicates the incorporated nucleotide, and the sequence is determined based on the order of nucleotide additions.
- Illumina sequencing offers several advantages, including high accuracy, scalability, and cost-effectiveness. It enables the generation of large amounts of sequence data in a single run, making it suitable for a wide range of applications such as whole-genome sequencing, targeted sequencing, and transcriptomics.
- The development of Illumina sequencing has revolutionized the field of genomics and has played a crucial role in advancing our understanding of genetics and molecular biology. Continued advancements and refinements in Illumina sequencing technology have further improved its performance and expanded its applications in various research areas.
Single molecule real time (RNAP) sequencing
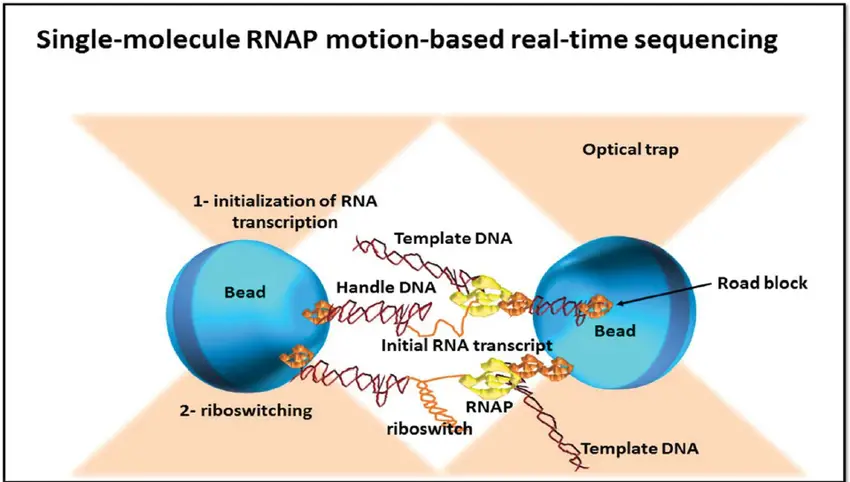
- Single molecule real-time (SMRT) sequencing, also known as RNAP sequencing, is a next-generation sequencing technology developed by Pacific Biosciences. It utilizes the activity of RNA polymerase (RNAP) to sequence DNA molecules in real-time.
- In SMRT sequencing, a single DNA molecule is immobilized on a surface, and a polymerase enzyme, typically an RNA polymerase, is attached to a polystyrene bead. The distal end of the sequenced DNA molecule is also attached to another bead. Both beads are placed in optical traps, which allow for the measurement of their relative distance.
- As the RNA polymerase moves along the DNA template during transcription, the beads are brought closer together or move apart, depending on the movement of the polymerase. These changes in the relative distance between the beads are recorded at a single nucleotide resolution using specialized optical detection systems.
- To determine the DNA sequence, the sequencing process involves repeated cycles of nucleotide incorporation. The four types of nucleotides, A, C, G, and T, are provided at lowered concentrations. When a nucleotide is incorporated by the polymerase, it emits a signal that is detected and recorded. By analyzing the time and intensity of these signals, the DNA sequence can be deduced.
- SMRT sequencing offers several advantages. It is a single-molecule sequencing technology, which means that individual DNA molecules are sequenced without the need for amplification. This reduces bias and errors introduced by PCR amplification. SMRT sequencing also has the capability to capture epigenetic modifications, such as DNA methylation, in real-time during sequencing.
- Although SMRT sequencing has relatively lower throughput compared to some other next-generation sequencing platforms, it provides long read lengths, making it suitable for applications such as de novo genome assembly, structural variant detection, and full-length transcript sequencing.
- In recent years, improvements in the SMRT sequencing technology have been made, enhancing the accuracy and read lengths, and expanding its applications in genomics and molecular biology research.
Single molecule SMRT sequencing
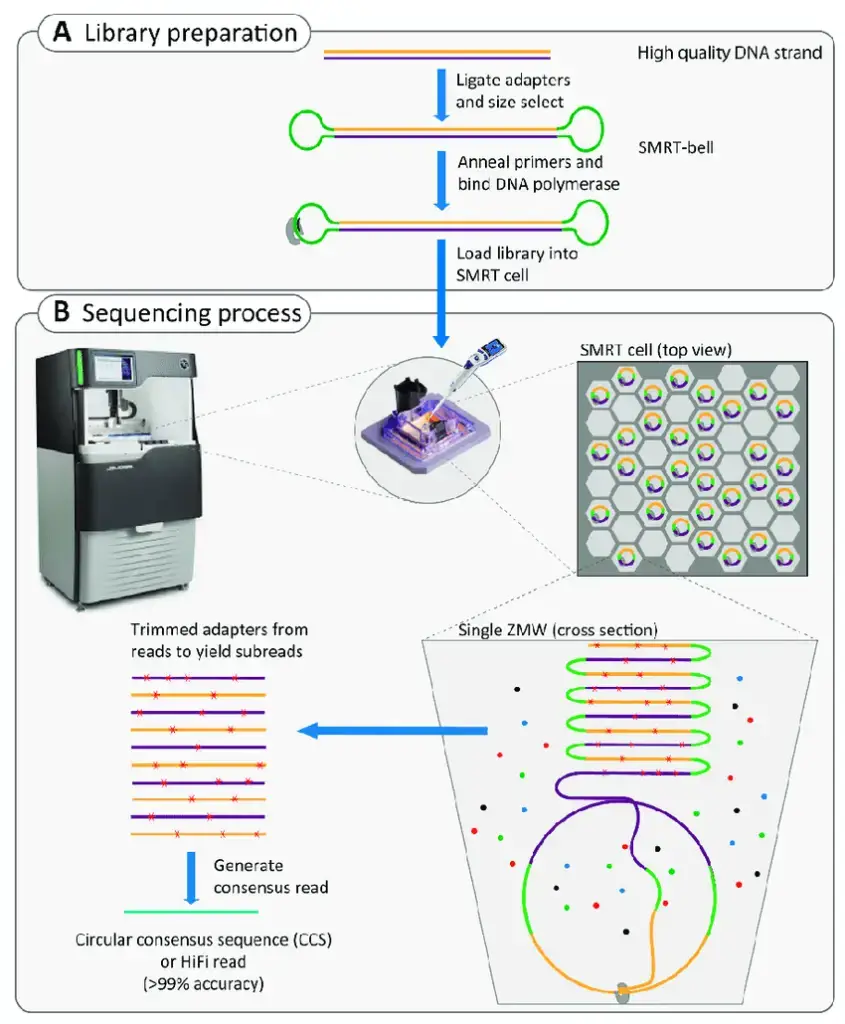
- Single molecule SMRT (Single Molecule Real-Time) sequencing is a revolutionary DNA sequencing technology developed by Pacific Biosciences. It is based on the concept of sequencing by synthesis and offers unique capabilities for high-throughput, long-read sequencing.
- In SMRT sequencing, DNA molecules are synthesized within specialized structures called zero-mode waveguides (ZMWs). These ZMWs are tiny well-like containers, each with a capturing tool located at the bottom. Only a single DNA molecule is present in each ZMW during sequencing.
- The sequencing process begins by introducing a DNA polymerase and fluorescently labeled nucleotides into the ZMWs. Unlike other sequencing methods, SMRT sequencing uses an unmodified polymerase and freely flowing fluorescent nucleotides in solution. As the DNA polymerase incorporates the nucleotides into the growing DNA strand, the fluorescence signal is emitted.
- The design of the ZMWs is critical in SMRT sequencing. They are constructed in such a way that only the fluorescence occurring near the bottom of the well is detected. This arrangement enables highly sensitive detection of the fluorescence signal, resulting in accurate base calling.
- After each nucleotide is incorporated, the fluorescent label is cleaved from the nucleotide, leaving behind an unmodified DNA strand. This allows for the detection of natural DNA modifications, such as methylation, as the polymerase kinetics are observed during sequencing.
- One of the notable advantages of SMRT sequencing is its ability to generate long reads. The technology enables reads of up to 1000 nucleotides, surpassing the read lengths of many other sequencing platforms. This makes SMRT sequencing particularly valuable for applications requiring the assembly of complex genomes, resolving structural variations, and studying epigenetic modifications.
- SMRT sequencing has significantly contributed to advancing our understanding of genomics and molecular biology. It has opened doors to explore previously inaccessible regions of the genome, unravel complex genetic variations, and decipher the epigenetic landscape of DNA molecules.
- As the technology continues to evolve and improve, single molecule SMRT sequencing holds great promise for diverse fields, including genomics research, personalized medicine, and precision agriculture. Its ability to provide long reads and detect nucleotide modifications offers unique insights into the complexity of the genome and its functional characteristics.
DNA nanoball sequencing
- DNA nanoball sequencing is a high-throughput sequencing technology that is utilized to determine the entire genomic sequence of an organism. It offers a cost-effective approach for sequencing a large number of DNA nanoballs in a single run.
- The sequencing process in DNA nanoball sequencing involves the use of rolling circle replication to amplify fragments of genomic DNA molecules. The DNA fragments are circularized and replicated multiple times to create DNA nanoballs, which are small, dense spheres composed of many replicated copies of the original DNA fragment.
- One of the advantages of DNA nanoball sequencing is its ability to sequence a large number of DNA nanoballs per run. This high throughput allows for efficient sequencing of genomes at a relatively low reagent cost compared to other next-generation sequencing platforms.
- However, a limitation of this technology is that only short sequences of DNA are determined from each DNA nanoball. This presents challenges when it comes to mapping the short reads to a reference genome, as longer sequences are generally more informative for accurate genome assembly and analysis. Nonetheless, with appropriate bioinformatics tools and algorithms, it is still possible to overcome this challenge and obtain meaningful genomic information from the short reads.
- DNA nanoball sequencing has been employed in various genome sequencing projects, contributing to the understanding of the genetic makeup of different organisms. Its cost-effectiveness and high throughput make it a valuable tool for large-scale sequencing endeavors.
- The technology continues to be developed and refined, and it is scheduled to be used in upcoming genome sequencing projects. With further advancements in sequencing technologies and bioinformatics tools, DNA nanoball sequencing holds promise for providing valuable genomic insights in the future.
Pyrosequencing
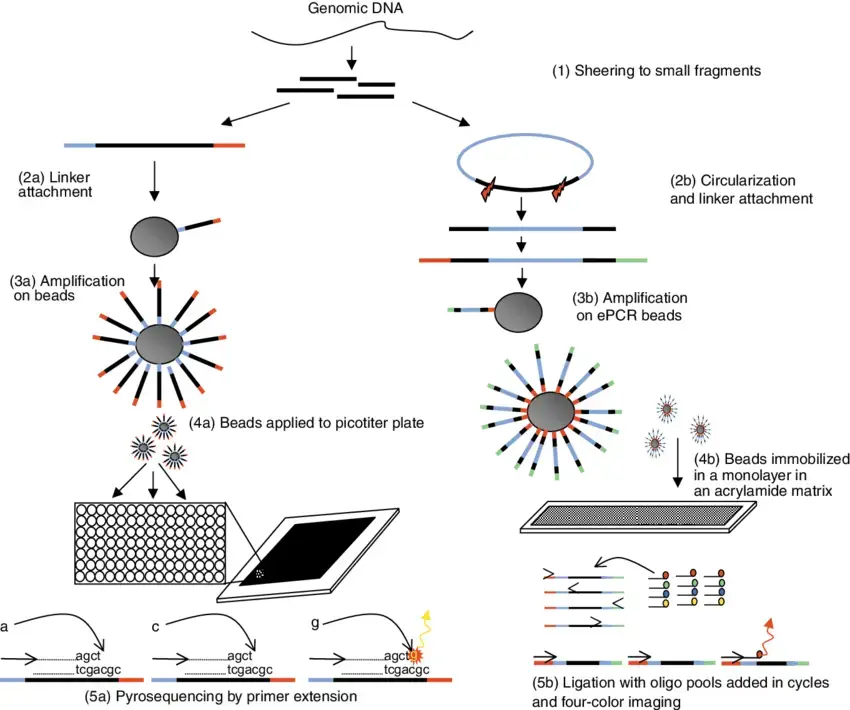
- Pyrosequencing is a sequencing method that was parallelized and developed by 454 Life Sciences, which is now a part of Roche Diagnostics. It offers an alternative approach to DNA sequencing and has its own unique characteristics.
- In pyrosequencing, the DNA amplification process takes place inside water droplets that are dispersed in an oil solution, a technique known as emulsion PCR. Each droplet contains a single DNA template that is attached to a primer-coated bead, resulting in the formation of clonal colonies. This clonal amplification allows for the amplification of individual DNA templates, enabling the sequencing of multiple DNA molecules in parallel.
- The sequencing machine used in pyrosequencing consists of numerous picolitre-volume wells, with each well containing a single bead and sequencing enzymes. The sequencing process involves the addition of individual nucleotides to the DNA template, and as each nucleotide is incorporated, a series of enzymatic reactions takes place. These reactions utilize luciferase, which generates light as a result of nucleotide incorporation.
- The emitted light is detected by the sequencing machine, and the intensity of the light corresponds to the identity of the added nucleotide. By detecting the light emitted during the sequencing process, pyrosequencing generates sequence read-outs, providing information about the DNA sequence.
- Pyrosequencing offers intermediate read length and cost per base compared to Sanger sequencing, on one end, and newer technologies like Solexa and SOLiD sequencing, on the other. While it may not provide the same read lengths and scalability as some of the more recent sequencing technologies, pyrosequencing offers a valuable balance between read length, cost, and throughput.
- Pyrosequencing has found applications in various fields, including genomics, genetics, and microbiology. It has contributed to the understanding of genetic variation, gene expression profiling, and the study of microbial communities.
- Although pyrosequencing has been largely replaced by newer sequencing technologies, it played a significant role in advancing the field of DNA sequencing and provided valuable insights into DNA analysis. Its development and utilization paved the way for the continued innovation and progress in the field of high-throughput sequencing.
Limitations of Next-Generation Sequencing (NGS)
Despite its numerous advantages, Next-Generation Sequencing (NGS) technologies also have some limitations that need to be considered. Here are a few key limitations:
- Infrastructure Requirements: Implementing NGS in the clinical setting requires significant infrastructure, including high computational capacity and storage resources. Managing and analyzing the vast amount of data generated by NGS requires specialized expertise and bioinformatics support. Establishing and maintaining such infrastructure can be costly and may require ongoing investments.
- Data Analysis and Interpretation: NGS produces massive amounts of data, and extracting clinically relevant information from this data can be a complex task. Skilled personnel are required to perform comprehensive data analysis and interpretation. The development of robust analytical pipelines and user-friendly interfaces is crucial for efficient and accurate interpretation of NGS results.
- Cost Considerations: While the actual cost of sequencing using NGS platforms has become increasingly affordable, the overall cost-effectiveness of NGS depends on several factors. Running large batches of samples is often necessary to make NGS economically viable, which may require centralization of sequencing facilities. Initial capital investments for equipment and infrastructure can be significant. However, once established, NGS facilities have the potential to offer economic benefits and improvements in patient care on a national scale.
- Standardization and Quality Control: Ensuring consistent and reliable results across different NGS platforms and laboratories can be challenging. Standardization of protocols, quality control measures, and data analysis pipelines is essential to minimize technical variability and ensure the accuracy and reproducibility of NGS results.
- Limited Read Lengths: Although NGS technologies have made significant progress in generating longer reads, the read lengths are still shorter compared to traditional Sanger sequencing. This can pose challenges in certain applications, such as de novo genome assembly and identification of structural variations in the genome.
- Detection of Structural Variants: While NGS is highly effective in detecting single nucleotide variations and small insertions/deletions, accurately identifying larger structural variants (e.g., duplications, inversions) can be more challenging. Specialized analysis methods and additional validation techniques are often required to detect and characterize these types of variants.
Despite these limitations, ongoing advancements in NGS technologies and bioinformatics tools continue to address many of these challenges. As the field evolves, it is expected that the limitations of NGS will be further overcome, leading to wider adoption and increased utility in clinical and research settings.
Applications of Next-Generation Sequencing (NGS)
Next-Generation Sequencing (NGS) has revolutionized various fields of research and healthcare due to its high-throughput and cost-effective nature. Here are some notable uses of NGS:
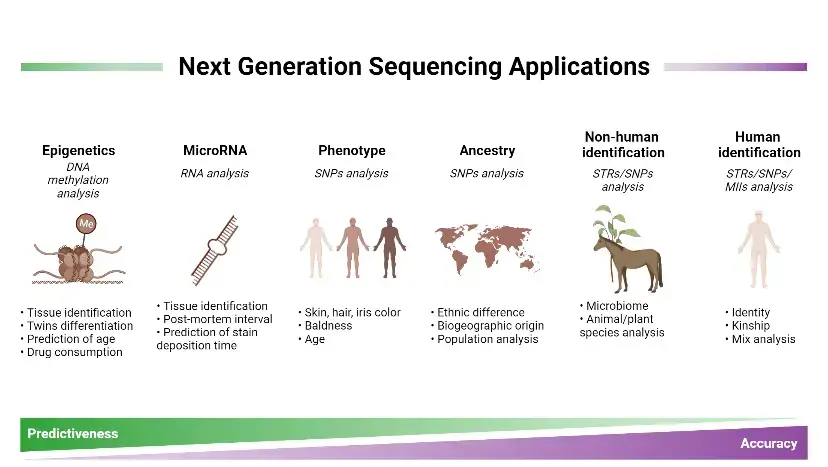
- Clinical Genetics: NGS has significantly impacted clinical genetics by enabling comprehensive analysis of the genome to improve patient care. It can capture a broader spectrum of mutations compared to traditional Sanger sequencing, allowing the identification of small base changes, insertions/deletions, large genomic deletions, and rearrangements. NGS provides a more complete view of genomic variation in a single experiment, reducing the need for multiple assays. It can aid in the diagnosis of unexplained syndromes, uncover novel disease-causing genes, and detect mosaic mutations with higher sensitivity.
- Microbiology: NGS has transformed microbiology by providing a genomic definition of pathogens. Instead of relying on conventional characterizations, such as morphology and staining properties, NGS allows for a comprehensive characterization of pathogens based on their genomes. This genomic information helps identify drug sensitivity, trace sources of infection outbreaks, and understand the relationship between different pathogens. NGS has been instrumental in tracking and managing outbreaks of infections, such as methicillin-resistant Staphylococcus aureus (MRSA) in healthcare settings.
- Oncology: NGS plays a crucial role in cancer genomics, where it allows for the systematic study of cancer genomes in their entirety. By analyzing the somatically acquired mutations in cancer cells, NGS provides a more precise diagnosis, classification, and prognosis of the disease. It also helps identify potential “druggable” mutations that can be targeted by mutation-specific drugs. NGS-based cancer sequencing offers the potential for personalized cancer management, enabling tailored treatment strategies for individual patients.
- Personalized Medicine: NGS is driving the development of personalized medicine approaches. By analyzing an individual’s genome, NGS enables the identification of genetic variants associated with disease susceptibility, drug response, and adverse reactions. This information can guide personalized treatment decisions, drug selection, and dosage optimization, leading to improved patient outcomes and reduced adverse effects.
- Infectious Disease Surveillance: NGS is transforming infectious disease surveillance by providing rapid and accurate identification of pathogens. By sequencing the genomes of infectious agents, NGS enables the tracking of disease outbreaks, monitoring the evolution of pathogens, and identifying drug resistance markers. This information is crucial for effective public health responses and the development of targeted interventions.
- Agricultural and Environmental Research: NGS has applications in agricultural and environmental research, allowing for the analysis of plant and animal genomes, microbiomes, and environmental DNA. It helps in understanding genetic diversity, studying evolutionary relationships, identifying beneficial traits, and monitoring environmental changes.
FAQ
Q1: What is Next-Generation Sequencing (NGS)?
A1: Next-Generation Sequencing, or NGS, is a high-throughput DNA sequencing technology that enables the rapid and simultaneous sequencing of millions to billions of DNA fragments.
Q2: How does NGS differ from traditional sequencing methods?
A2: NGS differs from traditional methods, such as Sanger sequencing, by its ability to generate massive amounts of sequencing data in a shorter time and at a lower cost per base.
Q3: What are the main applications of NGS?
A3: NGS has a wide range of applications, including whole genome sequencing, targeted sequencing, transcriptome analysis, metagenomics, epigenetics, and cancer genomics, among others.
Q4: How does NGS work?
A4: NGS works by fragmenting the DNA, attaching adapters to the fragments, and then amplifying and sequencing the fragments simultaneously. The resulting sequences are then assembled and analyzed using bioinformatics tools.
Q5: What are the advantages of NGS?
A5: NGS offers higher throughput, faster turnaround time, greater depth of coverage, and the ability to detect rare variants and complex genetic variations.
Q6: What are the limitations of NGS?
A6: Some limitations of NGS include the requirement for advanced bioinformatics analysis, the generation of large amounts of data that need to be managed and interpreted, and challenges in accurately mapping short reads to reference genomes.
Q7: What is the cost of NGS?
A7: The cost of NGS has significantly decreased over the years. It depends on factors such as the type of sequencing platform, the size of the genome or region being sequenced, and the desired coverage depth.
Q8: What are the major sequencing platforms used in NGS?
A8: Popular NGS platforms include Illumina (Solexa), Ion Torrent, Pacific Biosciences (PacBio), and Oxford Nanopore Technologies.
Q9: What is the typical read length in NGS?
A9: The read length in NGS varies depending on the sequencing platform and technology used. It can range from a few tens of base pairs up to several kilobases.
Q10: How is NGS advancing research and medicine?
A10: NGS has revolutionized genomics research and medical diagnostics by enabling comprehensive analysis of genomes, identification of disease-causing mutations, personalized medicine, and the discovery of novel genetic variants associated with diseases.
References
- Metzker, M. L. (2010). Sequencing technologies—The next generation. Nature Reviews Genetics, 11(1), 31–46.
- Mardis, E. R. (2013). Next-generation sequencing platforms. Annual Review of Analytical Chemistry, 6, 287–303.
- Goodwin, S., McPherson, J. D., & McCombie, W. R. (2016). Coming of age: Ten years of next-generation sequencing technologies. Nature Reviews Genetics, 17(6), 333–351.
- Kwok, P. Y. (2010). Methods for genotyping single nucleotide polymorphisms. Annual Review of Genomics and Human Genetics, 12, 221–237.
- Shendure, J., & Ji, H. (2008). Next-generation DNA sequencing. Nature Biotechnology, 26(10), 1135–1145.
- Metzker, M. L. (2009). Sequencing technologies—the next generation. Nature Reviews Genetics, 11(1), 31–46.
- Reuter, J. A., Spacek, D. V., & Snyder, M. P. (2015). High-throughput sequencing technologies. Molecular Cell, 58(4), 586–597.
- Mardis, E. R. (2017). DNA sequencing technologies: 2006–2016. Nature Protocols, 12(2), 213–218.
- Liu, L., Li, Y., Li, S., Hu, N., He, Y., Pong, R., … & Wang, J. (2012). Comparison of next-generation sequencing systems. Journal of Biomedicine and Biotechnology, 2012, 1–11.
- Loman, N. J., & Quinlan, A. R. (2014). Poretools: A toolkit for analyzing nanopore sequence data. Bioinformatics, 30(23), 3399–3401.
- Behjati S, Tarpey PS. What is next generation sequencing? Arch Dis Child Educ Pract Ed. 2013 Dec;98(6):236-8. doi: 10.1136/archdischild-2013-304340. Epub 2013 Aug 28. PMID: 23986538; PMCID: PMC3841808.
- https://www.thermofisher.com/in/en/home/life-science/sequencing/sequencing-learning-center/next-generation-sequencing-information/ngs-basics/what-is-next-generation-sequencing.html
- https://www.illumina.com/science/technology/next-generation-sequencing.html#:~:text=What%20is%20NGS%3F,regions%20of%20DNA%20or%20RNA.
Related Posts
- DNA Library – Types, Construction, Applications
- Emulsion PCR – Principle, Procedure, Advantages, Limitations, Uses
- TaqMan Probe – Definition, Principle, Applications
- Differences Between Sensitivity, Specificity, False positive, False negative
- Protein Synthesis (Translation)- Definition, Steps, Sites, Machinery