Contents
What is Northern blotting?
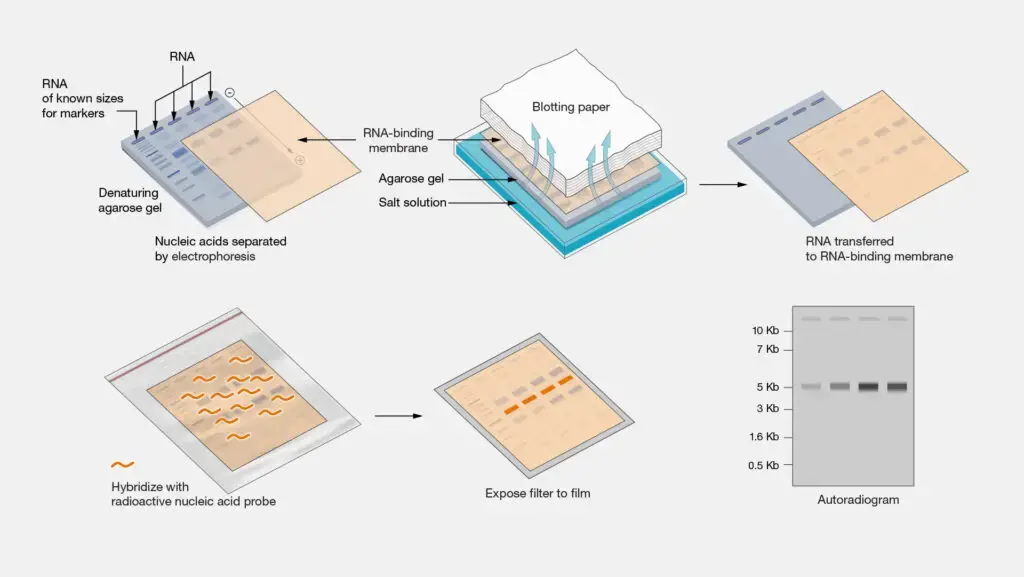
Northern blotting is a molecular biology technique used to detect and analyze RNA molecules. It involves the separation of RNA fragments based on their size through gel electrophoresis, followed by the transfer of the separated RNA molecules onto a solid membrane. The immobilized RNA fragments on the membrane are then hybridized with specific probes, typically complementary DNA (cDNA) or RNA molecules, that bind to the target RNA sequences. The probes are labeled with radioactive or non-radioactive tags, enabling the visualization and detection of the target RNA molecules. Northern blotting allows researchers to study gene expression patterns, mRNA abundance, alternative splicing, and other aspects of RNA biology. It has been a valuable tool in molecular biology research, contributing to our understanding of gene regulation, development, and disease mechanisms.
Northern blotting Protocol
- RNA Extraction: a. Collect the biological sample (e.g., tissue, cells) and extract total RNA using a suitable RNA extraction method (e.g., TRIzol, RNeasy kit). b. Ensure the RNA is of high quality and free from contaminants, such as DNA, proteins, and RNases. Quantify the RNA concentration using a spectrophotometer or fluorometer.
- RNA Denaturation and Gel Electrophoresis: a. Prepare a denaturing gel by mixing appropriate amounts of denaturing gel solution (e.g., formaldehyde-containing agarose gel) with RNA gel loading buffer. b. Add formaldehyde to the gel and heat the mixture to denature the RNA samples. c. Load the denatured RNA samples onto the gel wells, along with RNA size markers. d. Perform gel electrophoresis in a suitable running buffer at a constant voltage until the RNA fragments are well-separated according to their sizes.
- Transfer of RNA to a Membrane: a. Prepare a nylon or nitrocellulose membrane and cut it to the appropriate size for transfer. b. Soak the gel in a transfer buffer (e.g., 20x SSC) and assemble the transfer setup (e.g., sandwiching the gel between the membrane and absorbent materials). c. Apply gentle pressure or use capillary action to transfer the RNA from the gel onto the membrane over several hours to overnight.
- Membrane Pre-Hybridization: a. Pre-hybridize the membrane in a hybridization buffer (e.g., containing blocking agents such as Denhardt’s solution or BSA) to minimize non-specific binding. b. Incubate the membrane at an appropriate temperature (usually around 42°C) for 1-2 hours.
- Probe Preparation and Hybridization: a. Prepare the specific DNA or RNA probe by labeling it with a suitable radioactive or non-radioactive label (e.g., radioactive isotopes, biotin, or digoxigenin). b. Denature the labeled probe (e.g., by heating) and add it to the pre-hybridization buffer. c. Incubate the membrane with the labeled probe solution overnight at the appropriate hybridization temperature (typically around 42°C).
- Post-Hybridization Washing: a. Wash the membrane with a series of stringent wash buffers to remove unbound probe and reduce background noise. b. Start with low-stringency washes, gradually increasing the stringency with higher concentrations of salt or detergent. c. The number and duration of wash steps may vary depending on the probe and specific experimental conditions.
- Probe Detection: a. If using a radioactive probe, expose the membrane to an X-ray film or phosphorimaging screen to detect the radioactive signals. b. For non-radioactive probes, use appropriate detection methods such as chemiluminescence or fluorescence-based systems.
- Analysis and Interpretation: a. Develop the X-ray film or visualize the signals using suitable imaging systems. b. Analyze and interpret the results, including the presence, size, and abundance of the target RNA molecules.
What is Southern blotting?
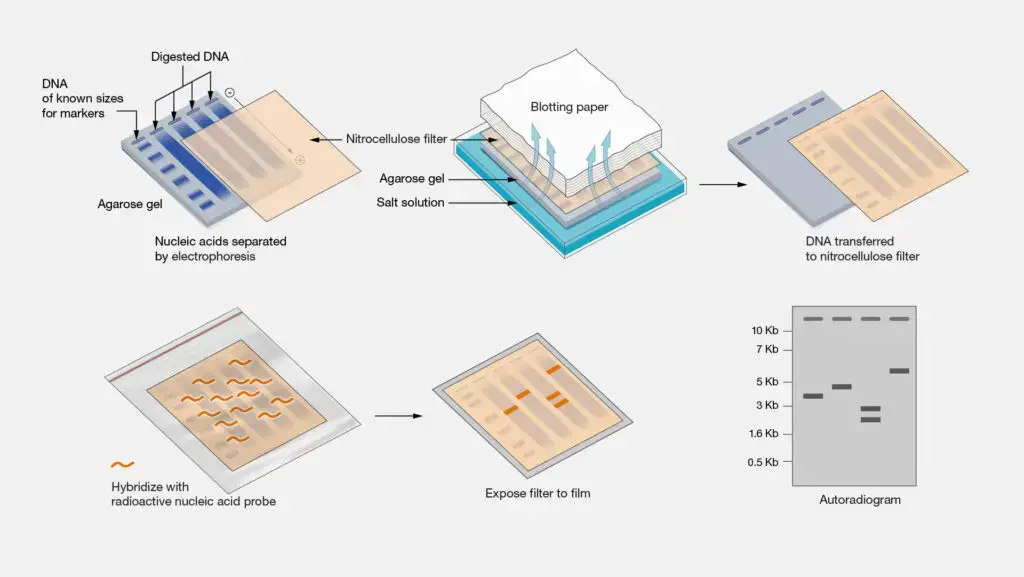
Southern blotting is a molecular biology technique used to detect and analyze DNA molecules. It was named after its inventor, Edwin Southern. The technique involves the separation of DNA fragments through gel electrophoresis, followed by the transfer of the separated DNA molecules onto a solid membrane, typically a nitrocellulose or nylon membrane. The immobilized DNA fragments on the membrane are then hybridized with specific DNA probes that are complementary to the target DNA sequences of interest. These probes are typically labeled with radioactive or non-radioactive tags, allowing the visualization and detection of the target DNA molecules. Southern blotting enables researchers to study DNA sequences, detect gene mutations, determine the presence or absence of specific DNA fragments, and analyze DNA methylation patterns. It has been widely used in various areas of molecular biology, including genetic research, disease diagnostics, and forensic analysis.
Southern blotting Protocol
- DNA Extraction: a. Collect the biological sample (e.g., tissue, cells) and extract genomic DNA using a suitable DNA extraction method (e.g., phenol-chloroform extraction, DNA purification kits). b. Ensure the DNA is of high quality, intact, and free from contaminants. Quantify the DNA concentration using a spectrophotometer.
- DNA Digestion and Gel Electrophoresis: a. Determine the appropriate restriction enzymes for DNA digestion based on the target sequences and experimental objectives. b. Digest the genomic DNA with the selected restriction enzymes in the appropriate reaction conditions. c. Prepare an agarose gel with suitable concentrations (typically 0.7-1.5%) and include DNA size markers. d. Load the digested DNA samples onto the gel wells. e. Perform gel electrophoresis in a suitable running buffer at a constant voltage until the DNA fragments are well-separated according to their sizes.
- DNA Denaturation and Transfer to a Membrane: a. Depurinate and denature the DNA in the gel by soaking it in a denaturation solution (e.g., alkali solution) and neutralization solution. b. Prepare a nylon or nitrocellulose membrane and cut it to the appropriate size for transfer. c. Assemble the transfer setup (e.g., sandwiching the gel between the membrane and absorbent materials) and transfer the DNA from the gel onto the membrane through capillary or vacuum transfer methods.
- Membrane Pre-Hybridization: a. Pre-hybridize the membrane in a hybridization buffer (e.g., containing blocking agents such as Denhardt’s solution or BSA) to minimize non-specific binding. b. Incubate the membrane at an appropriate temperature (usually around 42°C) for 1-2 hours.
- Probe Preparation and Hybridization: a. Prepare the specific DNA probe by labeling it with a suitable radioactive or non-radioactive label (e.g., radioactive isotopes, biotin, or digoxigenin). b. Denature the labeled probe (e.g., by heating) and add it to the pre-hybridization buffer. c. Incubate the membrane with the labeled probe solution overnight at the appropriate hybridization temperature (typically around 42°C).
- Post-Hybridization Washing: a. Wash the membrane with a series of stringent wash buffers to remove unbound probe and reduce background noise. b. Start with low-stringency washes, gradually increasing the stringency with higher concentrations of salt or detergent. c. The number and duration of wash steps may vary depending on the probe and specific experimental conditions.
- Probe Detection: a. If using a radioactive probe, expose the membrane to an X-ray film or phosphorimaging screen to detect the radioactive signals. b. For non-radioactive probes, use appropriate detection methods such as chemiluminescence or fluorescence-based systems.
- Analysis and Interpretation: a. Develop the X-ray film or visualize the signals using suitable imaging systems. b. Analyze and interpret the results, including the presence, size, and abundance of the target DNA fragments.
Northern blotting vs Southern blotting
| Feature | Northern Blotting | Southern Blotting |
|---|---|---|
| Target molecules | RNA | DNA |
| Purpose | Detect and analyze gene expression levels | Identify and characterize specific DNA sequences |
| Sample preparation | RNA extraction and purification required | DNA extraction and purification required |
| Denaturation | RNA is denatured with heat and/or formaldehyde | DNA is denatured with heat or alkali solution |
| Gel electrophoresis | Agarose gel is used | Agarose or polyacrylamide gel is used |
| Transfer method | Capillary or vacuum transfer | Capillary or vacuum transfer |
| Membrane type | Nitrocellulose or nylon membrane | Nitrocellulose or nylon membrane |
| Probe types | Radioactive or non-radioactive | Radioactive or non-radioactive |
| Probe labeling | Typically labeled with radioisotopes (e.g., 32P) | Labeled with radioisotopes (e.g., 32P) or non-radioactive |
| Hybridization | RNA probe hybridizes with complementary RNA sequences | DNA probe hybridizes with complementary DNA sequences |
| Detection | Autoradiography or chemiluminescence | Autoradiography or chemiluminescence |
| Sensitivity | Less sensitive than Southern blotting | More sensitive than Northern blotting |
| Specificity | Detects specific RNA sequences | Detects specific DNA sequences |
| Applications | Gene expression analysis, RNA quantification | DNA fragment analysis, DNA mapping, genetic testing |
| Target abundance | Can detect low-abundance RNA transcripts | Can detect low-abundance DNA sequences |
| RNA stability | RNA is less stable and prone to degradation | DNA is more stable and less prone to degradation |
| Reverse transcription | May involve reverse transcription step to convert RNA to DNA | Not applicable |
| Ribosomal RNA removal | Usually requires removal of ribosomal RNA | Not necessary for most applications |
| Complexity | Generally less complex than Southern blotting | Generally more complex than Northern blotting |
| Availability | Less commonly used compared to Southern blotting | More commonly used compared to Northern blotting |
Related Posts
- Difference Between Homologous Chromosomes and Sister Chromatids
- Difference between Monocarpic and Polycarpic Plants
- Differences Between Poisonous and Non-poisonous Snakes
- Differences Between Sensitivity, Specificity, False positive, False negative
- Anabolism vs Catabolism – Differences Between Anabolism and Catabolism