Contents
What is Intron?
- Introns are sequences that exist between two exons in eukaryotes. They do not code for proteins directly. They are eliminated prior to mRNA translation into proteins. Therefore, these introns are subjected to splicing.
- Introns, which are the non-coding portions of nucleotides, are not highly conserved. Therefore, removing introns is necessary to prevent the creation of erroneous proteins.
- Introns were found in 1977, when DNA sequencing was first introduced.
- Although it was known that mature eukaryotic mRNA molecules were shorter than the starting transcripts, it was believed that the ends of the transcripts were simply trimmed.
- When the two types of molecules were sequenced, it was discovered that this was not the case; the majority of the deleted transcript originated from core areas, not the extreme ends.
- This sparked substantial investigation into the removal of introns from transcripts and their potential function.
- It has been discovered that the frequency of introns inside distinct genomes varies greatly across the spectrum of living animals. Introns are relatively prevalent in the nuclear genomes of jawed vertebrates (such as humans and mice), where protein-coding genes almost always contain multiple introns, whereas introns are uncommon in the nuclear genes of certain eukaryotic microorganisms, such as baker’s/yeast. brewer’s (Saccharomyces cerevisiae).
- In contrast, the mitochondrial genomes of vertebrates are completely free of introns, but the mitochondrial genomes of eukaryotic bacteria may contain several introns.
- An extreme example is the Drosophila dhc7 gene, which contains a 3.6 megabase (Mb) intron and requires around three days to transcribe.
- The shortest known metazoan intron length, according to a 2015 study, is 30 base pairs (bp) and belongs to the human MST1L gene.
- The shortest known introns are found in heterotrich ciliates, such as Stentor coeruleus, where the majority (> 95%) are 15 or 16 bp in length.
Origin of Introns
- Evolution has utilised the presence of introns in genes to develop alternative splicing as a method for regulating gene expression.
- Exon shuffling has also made it possible to create new genes by mixing exons in different configurations. Several of these genes are present in mammals.
- However, it is less apparent where introns originate in the first place. Two hypotheses have been advanced.
- According to the first theory, the exon theory, introns were present in the progenote, the origin of all life forms, within the genes.
- These introns may have been capable of self-splicing. After the separation of the three major kingdoms, eubacteria and archaebacteria lost many of their introns.
- The alternative notion is that introns are a type of transposable element that can move into and out of genes that did not always have introns from the start.
- After the symbiotic event that generated mitochondria and chloroplasts, it would be feasible for group II introns to have infiltrated nuclear genes. This notion is supported by the fact that group II introns can hop into specified locations within genes.
- Both theories are somewhat supported by various types of observations, and both may be partially true.
Types of Introns
Group I introns, Group II introns, Nuclear pre-mRNA introns, and Transfer RNA itrons are the four types of introns.
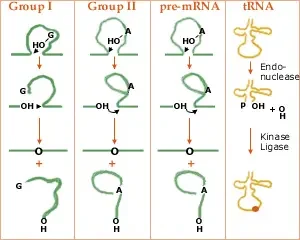
1. Group I introns
- They are huge ribozymes capable of catalysing their own excision from mRNA, tRNA, and rRNA precursors.
- The secondary structure of group I introns consists of a stem with nine loops, which is necessary for splicing.
- mRNA, tRNA, and rRNA from non-vertebrate mitochondria, chloroplasts, and nuclear genomes, bacteriophages, and eubacterial genomes all contain Group I introns.
- All group I introns fold in the same manner.
- Tomas Cech investigated group I introns and discovered that they can be removed in the absence of all proteins.
- This discovery gave rise to the concept of RNA enzymes and had a significant impact on our understanding of the evolution of life on Earth. Tomas Cech and Sidney Altman shared the 1989 Nobel Prize for discovering that another form of RNA, RNAse P, may catalyse the cleavage of tRNA precursors.
2. Group II Introns
- rRNA, tRNA, and mRNA also contain self-splicing ribozymes, although they cut themselves differently than group I introns.
- In addition, group II intron splicing requires the development of a lariat structure.
- Group II introns have been observed in cyanobacteria and proteobacteria as well as in mitochondria and chloroplasts.
- In addition, all group II introns share a similar folding pattern.
- Similar to introns of group I, introns of group II can self-splice. Proteins are required for both group I and group II intron splicing in vivo.
- It is probable that pre-mRNA introns and group II introns are evolutionarily connected due to the similarity between their excision mechanisms. If true, the snRNAs may have originated from introns of group II.
3. Spliceosomal introns/Nuclear pre-mRNA Introns
- They are present in the nucleus of protein-coding genes from which spliceosomes remove introns.
- Nuclear pre-mRNA introns (spliceosomal introns) are distinguished by the presence of unique intron sequences at the intron-exon borders.
- When splicing events are initiated, spliceosomal RNA molecules detect these sequences.
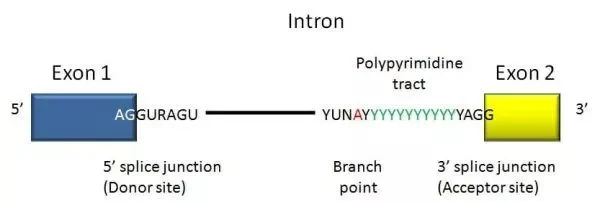
- In addition, they contain a branch point, which is a specific nucleotide sequence near the 3′ end of the intron that is covalently bonded to the 5′ end of the intron during the splicing process, resulting in a branched (lariat) intron.
- Other than these three short conserved components, the sequences of nuclear pre-mRNA introns are widely varied.
- Introns of nuclear pre-mRNA are frequently significantly longer than their surrounding exons.
4. Transfer RNA introns
- They are located in tRNA genes and must be eliminated by proteins (enzymes).
- The tRNA introns are unique in that they are removed by an enzyme that cuts the RNA, following which other enzymes phosphorylate (protein kinase) and religate the two tRNA halves.
Intron Structure
- In general, introns are significantly longer than exons; they can comprise up to 90 percent of a gene and can be over 10,000 nucleotides in length. Over 90% of human genes have introns, and the typical gene contains nine introns.
- A DNA segment that begins and ends with a particular sequence of nucleotides called an intron. As the boundary between introns and exons, these sequences are known as splice sites.
- The distinction between coding and non-coding DNA is essential for the development of functional genes.
- In humans and the vast majority of other vertebrates, introns start with 5′ GUA and terminate with 3′ CAG.
- Introns of both vertebrates and invertebrates contain additional conserved sequences, including a branch point involved in lariat (loop) creation.
Intron Function
- It has been demonstrated that introns likely play a crucial function in gene regulation and expression, despite the fact that they were once deemed “junk DNA” and to some extent still are.
- As introns lengthen genes, the possibility of crossing over and recombination between sister chromosomes is increased.
- This can result in novel gene variants via duplications, deletions, and exon shuffling. Additionally, introns permit alternate splicing.
- As the exons can be constructed in many ways, this permits a single gene to encode several proteins.
Splicing
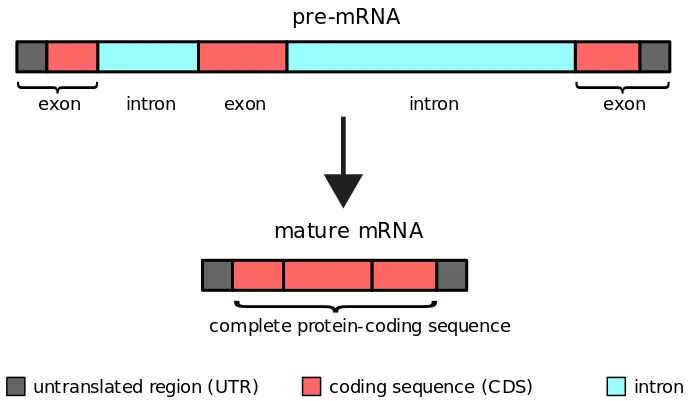
- RNA polymerase copies the complete gene, including introns and exons, into the initial mRNA transcript known as pre-mRNA or heterogeneous nuclear RNA during transcription (hrRNA).
- As introns are not transcribed, they must be eliminated prior to translation. Splicing is the process of removing introns and joining exons into a mature mRNA molecule, which occurs in the nucleus.
- Introns contain many splicing-related regions, including spliceosome recognition sites.
- These locations permit the spliceosome to identify the intron-exon boundary.
- The locations are identified by tiny nucleolar ribonucleoproteins (snRNPs). mRNA splicing involves a number of snRNPs that, when joined, form a spliceosome.
Steps of Splicing
Splicing occurs in three steps:
- At the 5′ terminus of the intron, cleavage of the phosphodiester link between the exon and the GU. One snRNP (U1) includes a sequence complementary to the 5′ splice site and binds there to induce splicing.
- A lariat or loop structure is formed. Near the 3′ end of the intron, the free 5′ end of the intron connects to a branch site, a conserved sequence. U2 binds to the branch site and recruits U1 to commence the lariat. A phosphodiester bond is then created between the free 5′ G and an A at the branch site to form the lariat.
- Cleavage of the phosphodiester link between the second exon and intron’s 3′ AG.
It is uncertain how the snRNPs and spliceosome determine which recognition sites to attach to, considering that introns can include thousands of base pairs and there are numerous cryptic splice sites where the recognition sequences are located elsewhere in the gene. Certain proteins (such as SR proteins), enhancers, and silencers are believed to be involved. Human diseases have also been linked to splicing silencers.
Alternative splicing
- In a process known as alternative splicing, introns and the splicing mechanism also allow for alternate gene products.
- Each discontinuous gene is composed of two or more exons, allowing for numerous exon assembly possibilities.
- Alternative splicing can generate anywhere between two and hundreds of distinct mRNAs.
- Alternative splicing is frequent in certain species but uncommon in others; it occurs in over 80% of human genes but only three Saccharomyces cerevisiae genes (yeast).
Steps of Alternative splicing
Alternative splicing can occur in a number of ways:
- Exon skipping: Exon skipping occurs when one (or more) exons are omitted from the final mRNA.
- Intron retention: a portion of the intron stays in the final mRNA due to improper splicing.
- Alternative splice site: The spliceosome eliminates a portion of one or more exons and the intron.
References
- https://www.genome.gov/genetics-glossary/Intron
- https://www.biologyonline.com/dictionary/intron
- https://en.wikipedia.org/wiki/Intron
- https://www.news-medical.net/life-sciences/What-are-introns-and-exons.aspx
- https://biologydictionary.net/intron/
- https://www.expii.com/t/what-are-introns-definition-role-in-transcription-10213
- https://educationalgames.nobelprize.org/educational/medicine/dna/a/splicing/introns.html
- https://www.aatbio.com/resources/faq-frequently-asked-questions/What-are-the-types-of-introns
- https://en.wikibooks.org/wiki/Structural_Biochemistry/Nucleic_Acid/RNA/RNA_modification/Introns#:~:text=There%20are%20four%20types%20of,splices%20itself%20out%20of%20genes.