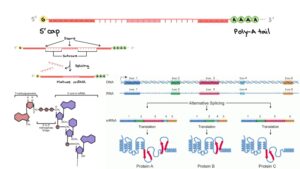
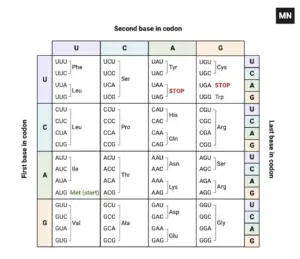
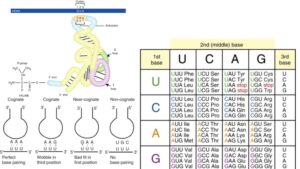
Many individuals believe that in the 1950s, American biologist James Watson and English physicist Francis Crick discovered DNA. In actuality, this is not true. DNA was discovered by the Swiss chemist Friedrich Miescher in the late 1860s. Then, in the decades that followed Miescher’s discovery, other scientists, notably Phoebus Levene and Erwin Chargaff, conducted a series of studies that revealed additional characteristics about the DNA molecule, such as its fundamental chemical components and the manner in which they were connected. Watson and Crick’s ground-breaking conclusion of 1953, that the DNA molecule exists in the form of a three-dimensional double helix, may never have been achieved without the scientific groundwork supplied by these pioneers.
The First Piece of the Puzzle: Miescher Discovered DNA
- In 1869, Swiss physiological chemist Friedrich Miescher discovered what he termed “nuclein” within the nuclei of human white blood cells, making 1869 a watershed year in genetic research. (The name “nuclein” was subsequently changed to “nucleic acid” and then to “deoxyribonucleic acid” or “DNA”)
- Miescher intended to identify and analyse not nuclein (which no one knew existed at the time) but the protein components of leukocytes (white blood cells).
- Miescher arranged for a local surgical clinic to bring him pus-coated, used patient bandages; once he received the bandages, he intended to wash them, filter out the leukocytes, then extract and identify the various proteins within the white blood cells.
- Miescher thought he had discovered a novel material when he identified a substance in the cell nucleus with chemical features unlike any protein, including a significantly higher phosphorous concentration and resistance to proteolysis (protein breakdown) (Dahm, 2008).
- Miescher, perceiving the significance of his results, penned, “It seems possible that a large family of such slightly different phosphorous-containing substances would develop as a set of nucleins that are analogous to proteins” (Wolf, 2003).
- The scientific community did not recognise the significance of Miescher’s discovery of nucleic acids for more than 50 years.
- Erwin Chargaff highlighted in a 1971 essay on the history of nucleic acid research that in a 1961 historical overview of nineteenth-century science, Charles Darwin was mentioned 31 times, Thomas Huxley was mentioned 14 times, but Miescher was not mentioned at all.
- Given that Miescher’s discovery of nucleic acids was unusual among the discoveries of the four primary biological components (i.e., proteins, lipids, polysaccharides, and nucleic acids) in that it could be “dated accurately… [to] one individual, one place, one date,” this exclusion is all the more surprising.
Laying the Groundwork: Levene Investigates the Structure of DNA
- Other scientists continued to research the molecular nature of the molecule once known as nuclein, even as Miescher’s name faded into obscurity in the twentieth century.
- The Russian biochemist Phoebus Levene was one of the other scientists. Over the course of his career, the physician-turned-chemist Levene published more than 700 articles on the chemistry of biological molecules. Levene is credited with numerous innovations.
- For example, he was the first to discover the order of the three major components of a single nucleotide (phosphate-sugar-base), the first to discover the carbohydrate component of RNA (ribose), the first to discover the carbohydrate component of DNA (deoxyribose), and the first to correctly identify the structure of RNA and DNA molecules.
- Prior to the discovery of the sugar-phosphate backbone of the DNA molecule, neither Levene nor any other scientist at the time knew how the individual nucleotide components of DNA were ordered in space.
- Due to the huge number of chemical groups made available for binding by each nucleotide component, there were countless possible combinations of the components.
- Several scientists proposed potential explanations for this phenomenon, but Levene’s “polynucleotide” hypothesis proved to be accurate. Levene claimed that nucleic acids are formed of a series of nucleotides, and that each nucleotide is composed of just one of four nitrogen-containing bases, a sugar molecule, and a phosphate group, based on years of hydrolysis-based research on yeast nucleic acids.
- Levene made his first proposal in 1919, dismissing previous hypotheses regarding the structure of nucleic acids. According to Levene, “Fresh facts and new evidence may cause its modification, but the polynucleotide structure of the yeast nucleic acid is beyond question” (1919).
- Indeed, the emergence of several new facts and an abundance of new data led to the modification of Levene’s theory. The ordering of nucleotides was a significant finding made during this time period.
- Levene devised a tetranucleotide structure in which the nucleotides are always linked in the same order (i.e., G-C-T-A-G-C-T-A and so on).
- Scientists subsequently found that Levene’s hypothesised structure for tetranucleotides was extremely simplistic and that the order of nucleotides along a stretch of DNA (or RNA) is, in reality, very changeable.
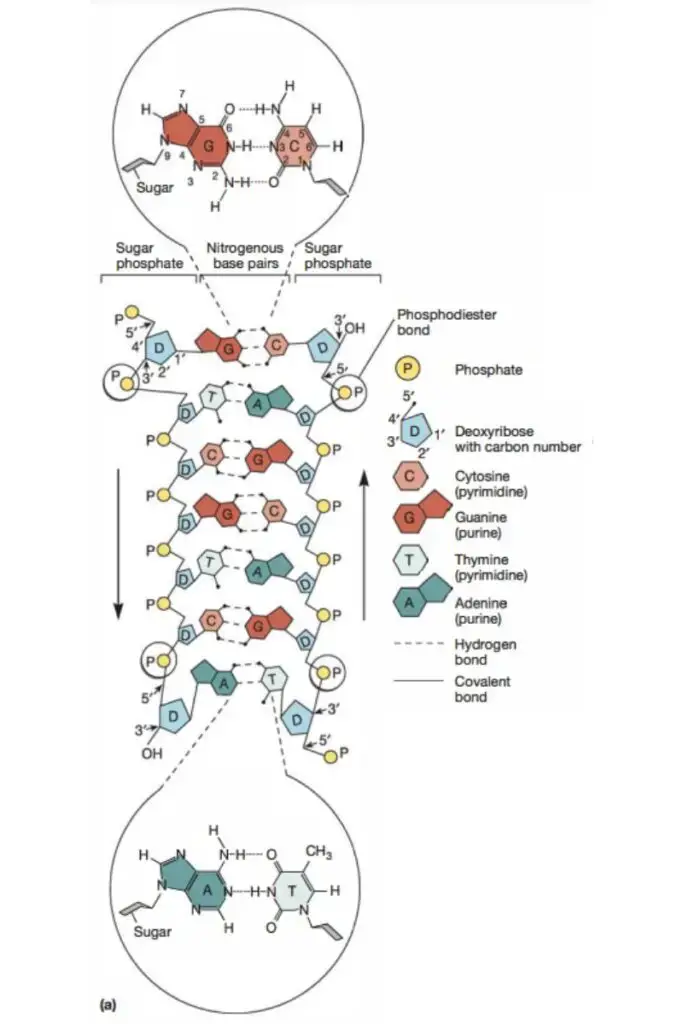
- In many respects, Levene’s hypothesised polynucleotide structure was true despite this revelation. We now know, for instance, that DNA is formed of a series of nucleotides and that each nucleotide has three components: a phosphate group; either a ribose (in the case of RNA) or deoxyribose (in the case of DNA) sugar; and a single nitrogen-containing base.
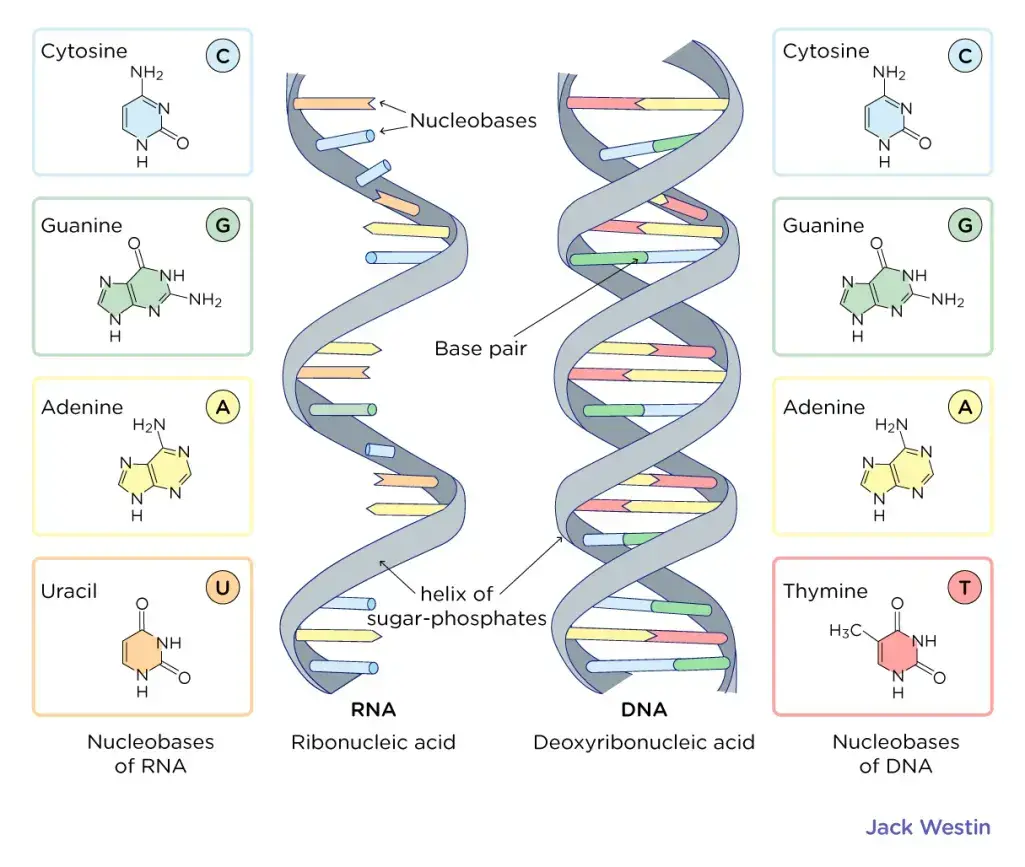
- We also know that there are two primary classes of nitrogenous bases: purines (adenine [A] and guanine [G]), each with two fused rings, and pyrimidines (cytosine [C], thymine [T], and uracil [U], each with a single ring). In addition, it is now generally acknowledged that RNA includes just A, G, C, and U (no T), whereas DNA has only A, G, and T. (no U).
Griffith’s Transformation Experiment
In 1928, Frederick Griffith, a pioneering British bacteriologist, conducted an experiment that fundamentally altered our understanding of genetics and bacterial pathology. His research, known as Griffith’s Transformation Experiment, provided crucial evidence for the role of genetic material in bacterial transformation.
Background and Initial Observations
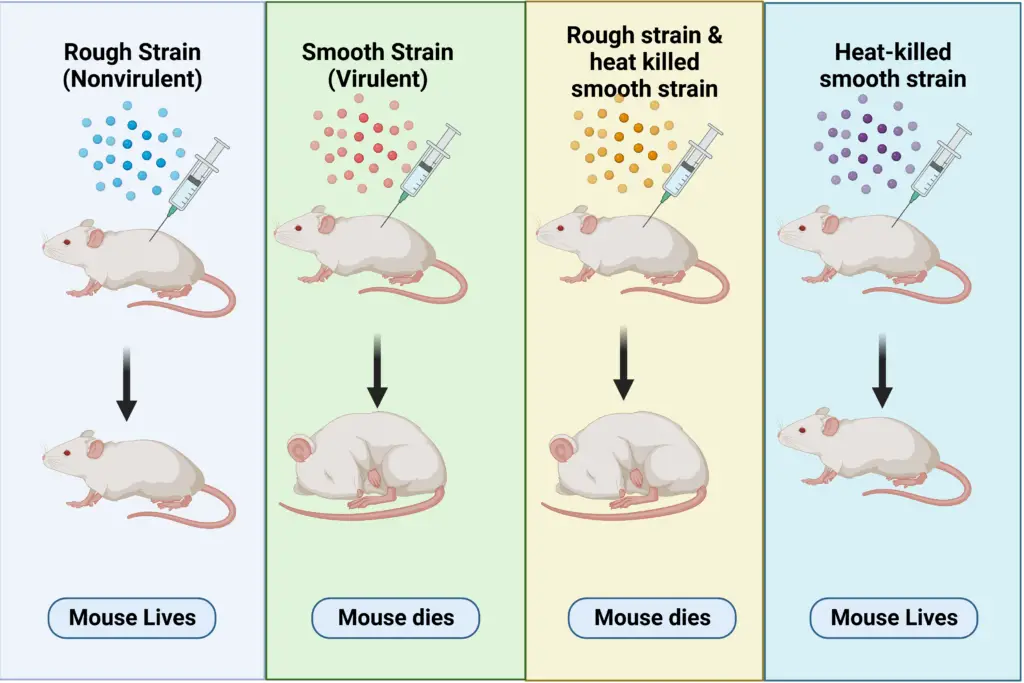
Griffith’s investigation was motivated by the high mortality rates associated with pneumococcal infections during the 1918 Spanish influenza pandemic. Streptococcus pneumoniae, the bacterium responsible for these infections, prompted Griffith to seek a method for vaccine development. During his research, he observed that S. pneumoniae existed in multiple strains, which were distinguishable based on their pathogenic properties. Specifically, he noted that:
- Type I S-Strain: These bacteria formed smooth, glistening colonies on agar plates. This smooth appearance was due to a protective polysaccharide capsule that shielded them from the host’s immune response. Consequently, the S-strain was virulent, causing severe pneumonia and death when injected into mice.
- Type II R-Strain: In contrast, these bacteria produced rough colonies and lacked the polysaccharide coat. They were non-virulent and did not cause disease when injected into mice, demonstrating their inability to evade the immune system.
Experimental Procedure and Results
Griffith’s pivotal experiment involved several key steps:
- Heat-Killed S-Strain Injection: Griffith injected mice with heat-killed S-strain bacteria. These bacteria had been subjected to high temperatures, rendering them non-viable. As expected, the mice survived, indicating that the heat-killed S-strain did not cause disease.
- Mixing Live R-Strain with Heat-Killed S-Strain: Griffith then mixed live R-strain bacteria with the heat-killed S-strain bacteria and injected this mixture into mice. Contrary to expectations, the mice developed pneumonia and died.
- Isolation of Bacteria from Dead Mice: Griffith isolated bacteria from the blood of the deceased mice and discovered that the bacteria were of the S-strain type. This was unexpected, as the R-strain used in the experiment was non-virulent.
Griffith’s results led him to hypothesize that some “transforming principle” from the dead S-strain bacteria had been transferred to the live R-strain bacteria. This transfer enabled the previously non-virulent R-strain bacteria to produce the protective polysaccharide capsule and exhibit virulence similar to the S-strain. Thus, the heat-killed S-strain bacteria had somehow converted the live R-strain bacteria into a virulent form.
This transforming principle, as identified by Griffith, laid the groundwork for the later discovery that DNA is the hereditary material responsible for genetic transformation. Although Griffith did not identify the chemical nature of the transforming principle, his work demonstrated that genetic information could be transferred between organisms, altering their characteristics.
Legacy and Impact
Griffith’s experiment was a cornerstone in the field of genetics and molecular biology, leading to further research by other scientists such as Oswald Avery, Colin MacLeod, and Maclyn McCarty, who ultimately identified DNA as the genetic material. Despite Griffith’s untimely death in 1941 due to a bomb during World War II, his pioneering work remains a fundamental contribution to our understanding of genetic transformation.
Strengthening the Foundation: Chargaff Formulates His “Rules”
- Erwin Chargaff was one of a handful of scientists that improved on Levene’s work by revealing further insights about the structure of DNA, opening the path for Watson and Crick.
- Austrian biochemist Chargaff had read the 1944 study by Oswald Avery and his colleagues at Rockefeller University that established that hereditary units, or genes, are made up of DNA.
- This article had a great effect on Chargaff, prompting him to initiate a study programme centred on the chemistry of nucleic acids. Regarding the findings of Avery, Chargaff (1971) wrote the following: “Almost abruptly, this discovery appeared to predict a chemistry of heredity and also made the nucleic acid nature of the gene probable… Avery provided us with the first text of a new language, or more rather, he showed us where to get it. I decided to look for this text.”
- Chargaff’s initial step in this investigation was to determine whether there were DNA differences between species. Chargaff drew two important conclusions after creating a new paper chromatography method for separating and identifying minute amounts of organic substances (Chargaff, 1950).
- Initially, he mentioned that the nucleotide content of DNA differs between species. In other words, as postulated by Levene, identical nucleotides do not recur in the same order.
- Second, Chargaff found that nearly all DNA, regardless of the organism or tissue type from which it originates, retains certain qualities despite variations in its composition.
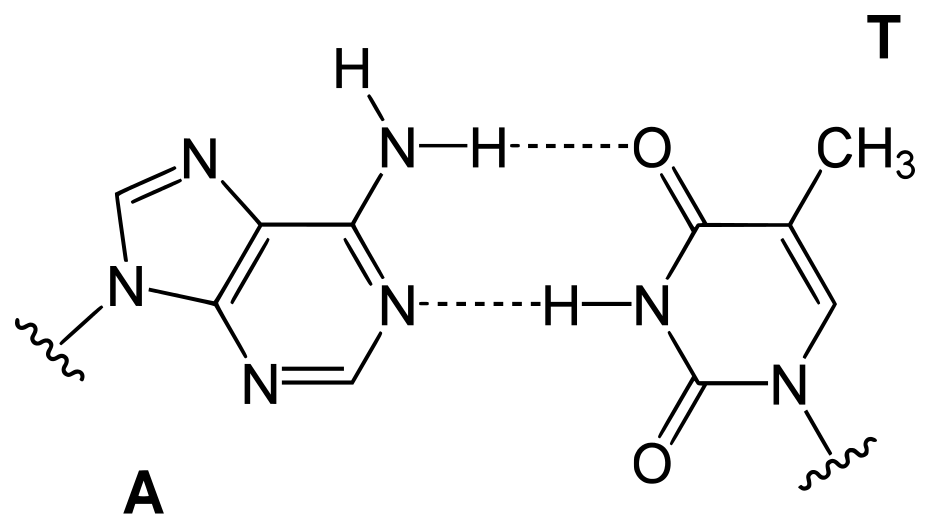
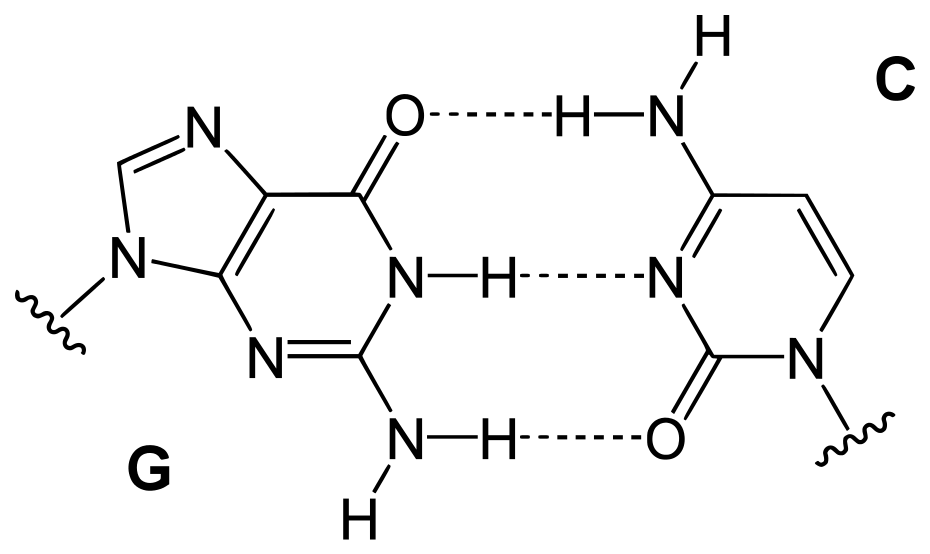
- In particular, the quantity of adenine (A) is typically comparable to the quantity of thymine (T), and the quantity of guanine (G) is typically comparable to the quantity of cytosine (C) (C).
- In other words, the total amount of purines (A + G) and the total amount of pyrimidines (C + T) are typically very close to one another. (This second significant finding is now referred to as “Chargaff’s rule.”)
- Chargaff’s discovery was essential to the later work of Watson and Crick, although Chargaff himself was unable to envision the explanation for these correlations, namely that A bound to T and C bound to G within the DNA molecular structure.
Avery, McCarty, and MacLeod Experiment
In 1943, Oswald Avery, Maclyn McCarty, and Colin MacLeod conducted a groundbreaking experiment at Rockefeller University that provided definitive evidence for DNA as the genetic material. Building upon Frederick Griffith’s earlier work, their research sought to identify the biochemical nature of the “transforming principle” Griffith had observed. This experiment was pivotal in advancing our understanding of genetic material.
Experimental Design and Methodology
Avery, McCarty, and MacLeod aimed to elucidate the transforming principle by isolating and analyzing its components. Their approach involved the following steps:
- Preparation of Cell Extracts:
- They began by heat-killing type III S-strain S. pneumoniae cells to ensure that the bacteria were non-viable.
- The cell extracts were then subjected to treatments to remove non-DNA components, specifically lipids and carbohydrates.
- Enzymatic Digestion:
- The extracts were treated with various enzymes to degrade specific macromolecules:
- RNases: Enzymes that degrade RNA.
- Proteases: Enzymes that break down proteins.
- DNases: Enzymes that digest DNA.
- Each type of enzyme was applied in separate tubes to determine the impact on the transforming principle.
- The extracts were treated with various enzymes to degrade specific macromolecules:
- Transformation Assays:
- Live type II R-strain cells were mixed with the treated heat-killed type III S-strain extracts and then introduced onto a culture medium.
- The culture medium contained antibodies specific to the type II R-strain, which helped distinguish between transformed and non-transformed bacteria by inactivating some of the R-strain cells.
Observations and Results
The results from these experiments were pivotal:
- DNase Treatment: In cultures where DNase was used, transformation did not occur. This result indicated that DNA, not RNA or proteins, was required for the transformation process.
- RNase and Protease Treatments: In cultures treated with RNases and proteases, transformation still occurred, reinforcing the conclusion that RNA and proteins were not the transforming principle.
Hershey and Chase Experiment
In 1952, Alfred Hershey and Martha Chase conducted a pivotal experiment that provided definitive evidence for DNA as the genetic material. Their work built upon the findings of Avery, McCarty, and MacLeod, and addressed lingering doubts about whether DNA or protein was the material responsible for genetic inheritance. Hershey and Chase utilized bacteriophages—viruses that infect bacteria—to explore this question.
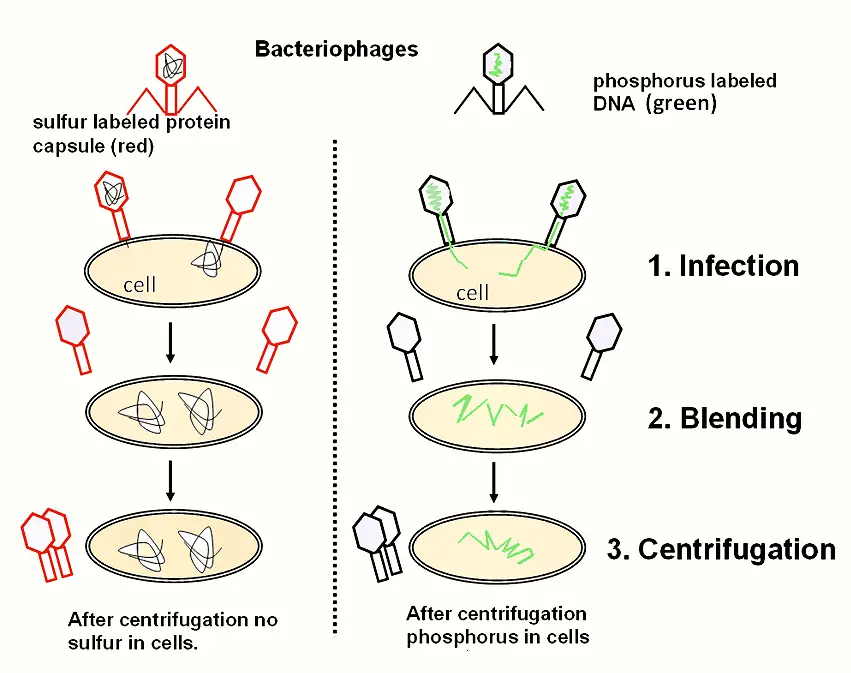
Experimental Design and Methodology
The Hershey and Chase experiment employed the T2 bacteriophage, a virus that specifically infects Escherichia coli (E. coli) bacteria. The key components of their experimental approach included:
- Preparation of Radioactive Phages:
- Phosphorus-32 (32P) Labeling: Phages were grown in a medium containing radioactive phosphorus-32. DNA, which contains phosphorus but not sulfur, was thus labeled with 32P.
- Sulfur-35 (35S) Labeling: Phages were also grown in a medium containing radioactive sulfur-35. Proteins, which contain sulfur but not phosphorus, were labeled with 35S.
- Infection Process:
- Radioactively labeled phages were used to infect E. coli cells. This allowed Hershey and Chase to trace which component of the phage—DNA or protein—entered the bacterial cells during infection.
- Separation of Phage and Bacterial Components:
- After allowing sufficient time for infection, the mixture was processed using a blender to remove phage particles that remained outside the bacterial cells.
- The blended mixture was then centrifuged. This process separated the heavier bacterial cells, which formed a pellet at the bottom of the centrifuge tube, from the lighter phage particles and other debris, which remained in the supernatant.
Observations and Results
The results of the Hershey and Chase experiment provided clear insights:
- Phosphorus-32 (32P) Distribution: The pellet, which contained the bacterial cells, was found to be radioactive with 32P. This indicated that the DNA, labeled with phosphorus, had entered the bacterial cells.
- Sulfur-35 (35S) Distribution: The supernatant, which contained the phage debris, was radioactive with 35S. This demonstrated that the proteins, labeled with sulfur, remained outside the bacterial cells.
Putting the Evidence Together: Watson and Crick Propose the Double Helix
- Chargaff’s discovery that A = T and C = G, together with Rosalind Franklin and Maurice Wilkins’ key X-ray crystallography work, contributed to Watson and Crick’s development of the three-dimensional, double-helical model for the structure of DNA.
- Recent developments in model building, or the creation of hypothetical three-dimensional structures based on known chemical distances and bond angles, a technique developed by American biologist Linus Pauling, also contributed to Watson and Crick’s finding.
- In reality, Watson and Crick feared being “scooped” by Pauling, who suggested an alternative hypothesis for the three-dimensional structure of DNA months before they did. However, Pauling’s forecast was ultimately inaccurate.
- Watson and Crick rearranged cardboard cutouts representing the different chemical components of the four bases and other nucleotide subunits on their desks, as though putting together a jigsaw.
- They were initially misled by an incorrect understanding of the configuration of the individual components in thymine and guanine (particularly the carbon, nitrogen, hydrogen, and oxygen rings).
- Watson decided to build new cardboard cutouts of the two bases only at the suggestion of American physicist Jerry Donohue, to determine whether a different atomic configuration may make a difference. It did.
- Not only did each pair of complimentary bases now fit together flawlessly (i.e., A with T and C with G), but the structure also reflected Chargaff’s rule.
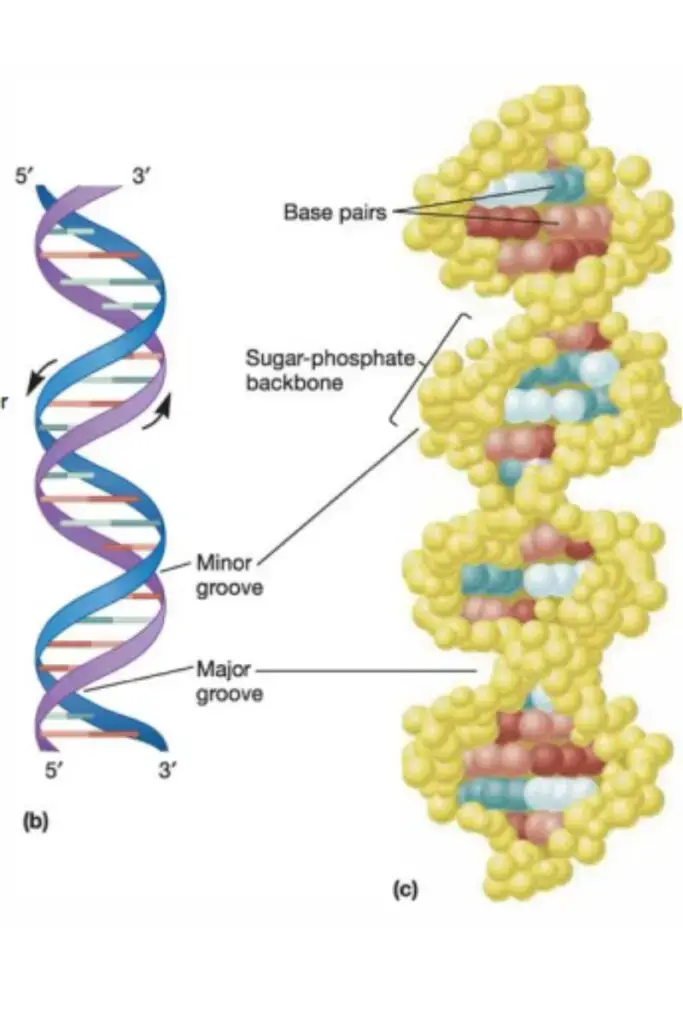
Although scientists have made slight modifications to the Watson and Crick model or expanded upon it since its conception in 1953, the model’s four basic characteristics remain unchanged. These characteristics are as follows:
- DNA is a double-stranded helix with hydrogen bonds connecting the two strands. A bases are always paired with Ts, and C bases are always paired with Gs, which is compatible with Chargaff’s rule and explains for it.
- The majority of DNA double helices are right-handed, meaning that if you held out your right hand with your thumb pointing upward and your fingers curled around your thumb, your thumb would represent the axis of the helix and your fingers would represent the sugar-phosphate backbone. Z-DNA is the only kind of DNA that is left-handed.
- The DNA double helix is anti-parallel, meaning that each strand’s 5′ end is coupled with the 3′ end of its corresponding strand (and vice versa). As depicted in Figure 4, nucleotides are connected by their phosphate groups, which bind the 3′ end of one sugar to the 5′ end of the following sugar.
- Not only are the DNA base pairs joined by hydrogen bonding, but the nitrogen-containing bases’ outer edges are exposed and potentially available for hydrogen bonding as well. These hydrogen bonds offer simple access to the DNA for other molecules, including proteins with crucial roles in DNA replication and expression.
The detection of three distinct conformations of the DNA double helix is one way in which scientists have elaborated on Watson and Crick’s concept. Thus, the exact geometries and dimensions of the double helix are variable. B-DNA is the type of DNA seen in the majority of live cells (it is portrayed in most schematics of the double helix and was postulated by Watson and Crick). Two more conformations exist: A-DNA, a shorter and wider form observed in dried DNA samples and seldom under normal physiological conditions, and Z-DNA, a left-handed conformation. Z-DNA is a kind of DNA that exists only sporadically in response to specific types of biological activity (Figure 5). Z-DNA was first found in 1979, but until recently its presence was largely disregarded. Since then, scientists have shown that some proteins tightly link to Z-DNA, suggesting that Z-DNA plays a vital biological role in viral illness defence (Rich & Zhang, 2003).
- https://old-ib.bioninja.com.au/higher-level/topic-7-nucleic-acids/71-dna-structure-and-replic/dna-experiments.html
- https://iubmb.onlinelibrary.wiley.com/doi/full/10.1002/bmb.20309
- http://www.nature.com/scitable/topicpage/discovery-of-dna-as-the-hereditary-material-340
- https://bio.libretexts.org/Bookshelves/Genetics/Online_Open_Genetics_(Nickle_and_Barrette-Ng)/01%3A_Overview_DNA_and_Genes/1.02%3A_DNA_is_the_Genetic_Material
- https://www.slideshare.net/slideshow/dna-as-genetic-material-237066240/237066240
- https://ocw.mit.edu/courses/7-01sc-fundamentals-of-biology-fall-2011/pages/molecular-biology/dna-structure-classic-experiments/
- https://www.yourgenome.org/theme/the-discovery-of-dna-the-molecule-of-life/
- https://www.sciencebuddies.org/science-fair-projects/science-projects/experiment-with-dna