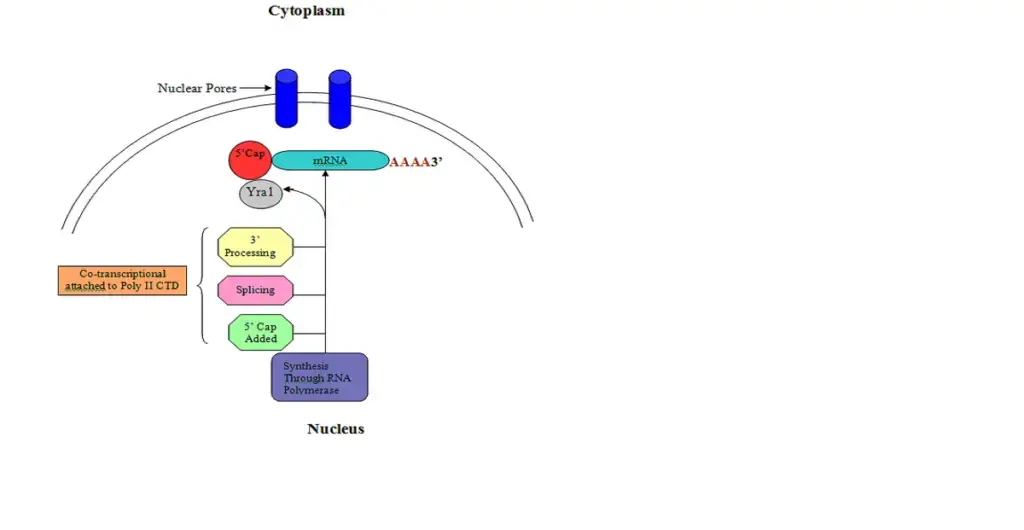
- In eukaryotes, transcription and translation occur in the nucleus and cytoplasm, respectively.
- In prokaryotes, transcription and translation of mRNA occur concurrently. Modifications to mRNA processing are minimal or nonexistent.
- In contrast, post-synthesis processing of pre-tRNA and pre-rRNA includes cleavage, addition of nucleotides, and chemical modification.
- Before being transported to the cytoplasm where they are translated by ribosomes, eukaryotic pre-mRNA undergoes 5′ capping, 3 cleavage/polyadenylation, splicing, and RNA editing.

What is 5′ capping?
The unique cap structure of eukaryotic mRNA is composed of 7-methylguanosine residues connected by a 5′-5′ triphosphate bridge.
- The five-prime cap (5′ cap) is a modified nucleotide at the 5′ end of various primary transcripts, such as precursor messenger RNA, in molecular biology.
- This process, known as mRNA capping, is tightly regulated and essential for the production of stable and mature messenger RNA capable of translation during protein synthesis.
- Both mitochondrial mRNA and chloroplastic mRNA lack a cap structure.
Structure of 5′ cap
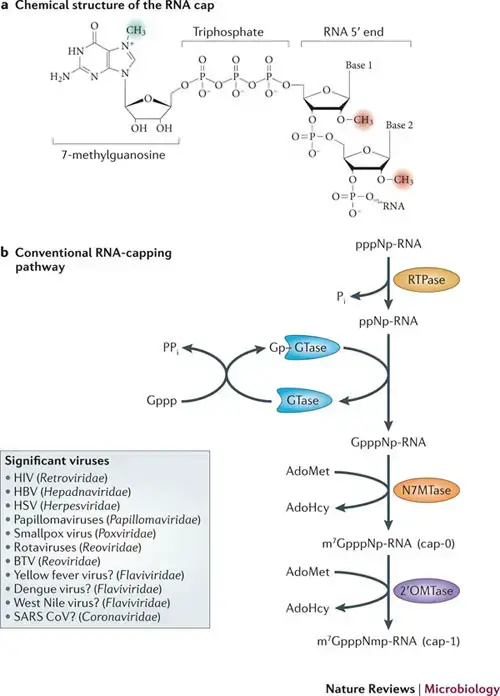
- The 5′ cap (cap-0) at the 5′ end of an mRNA molecule in eukaryotes consists of a guanine nucleotide linked to the mRNA through an unique 5′ to 5′ triphosphate bond.
- A methyltransferase methylates this guanosine at position 7 immediately after capping in vivo.
- It is known as a 7-methylguanylate cap, or m7G for short.
- Further alterations exist in multicellular eukaryotes and certain viruses, including the methylation of the 2′ hydroxy-groups of the first two ribose sugars at the 5′ end of the mRNA.
- Unlike cap-2, which has methylation 2′-hydroxy groups on the first two ribose sugars, cap-1 has a methylated 2′-hydroxy group on the first ribose sugar.
- The 5′ cap is chemically equivalent to the 3′ end of an RNA molecule (the 5′ carbon of the cap ribose is bonded, whereas the 3′ carbon is unbonded). This significantly increases resistance to 5′ exonucleases.
- Small nuclear RNAs feature distinct 5′-caps. Lsm-class snRNAs have 5′-monomethylphosphate caps, whereas Sm-class snRNAs have 5′-trimethylguanosine caps.
- Some RNAs in bacteria and maybe in higher species are capped with NAD+, NADH, or 3′-dephospho-coenzyme A.
- mRNA molecules can be decapped in all species through a process known as messenger RNA decapping.
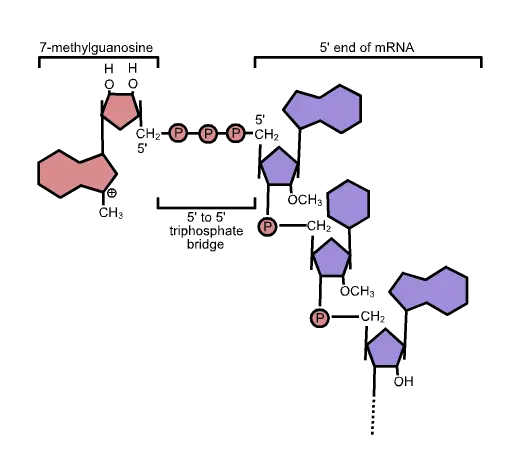
5’ Capping process
The starting point for capping with 7-methylguanylate is the unmodified, triphosphate-terminated 5′ end of an RNA molecule. Following the last nucleotide are three phosphate groups connected to the 5′ carbon. As nascent pre-mRNA is being generated, the capping procedure is commenced prior to the conclusion of transcription.
- RNA triphosphatase removes one of the terminal phosphate groups, leaving a bisphosphate group (i.e. 5′(ppN)[pN]n);
- mRNA guanylyltransferase adds GTP to the terminal bisphosphate while losing a pyrophosphate from the GTP substrate. This results in the formation of the 5′–5′ triphosphate linkage, 5′(Gp)(ppN)[pN]n;
- mRNA (guanine-N7-)-methyltransferase methylates the 7-nitrogen of guanine, and S-adenosyl-L-methionine is demethylated to create S-adenosyl-L-homocysteine, resulting in 5′(m7Gp)(ppN)[pN]n (cap-0);
- Normally, cap-adjacent alterations occur on the first and second nucleotides, yielding 5′(m7Gp)(ppN*)(pN*)[pN]n (cap-1 and cap-2);
- If the cap-adjacent nucleotide is 2′-O-ribose methyl-adenosine (i.e. 5′(m7Gp)(ppAm)[pN]n), it can be methylated at the N6 methyl site to create N6-methyladenosine, yielding 5′(m7Gp)(ppm6Am)[pN]n.
The capping mechanisms for NAD+, NADH, and 3′-dephospho-coenzyme A are distinct. Capping with NAD+, NADH, or 3′-dephospho-coenzyme A is achieved via a “ab initio capping mechanism” in which NAD+, NADH, or 3′-desphospho-coenzyme A serves as a “non-canonical initiating nucleotide” (NCIN) for transcription initiation by RNA polymerase and is thus directly incorporated into the RNA product. This “ab initio capping technique” can be implemented by both bacterial RNA polymerase and eukaryotic RNA polymerase II.
Function of 5’ Capping
- Control of nuclear exports
- Preventing degradative effects of exonucleases.
- Promotion of language (see ribosome and translation).
- Encouragement of 5′ proximal intron removal.
What is Poly(A) Polyadenylation?

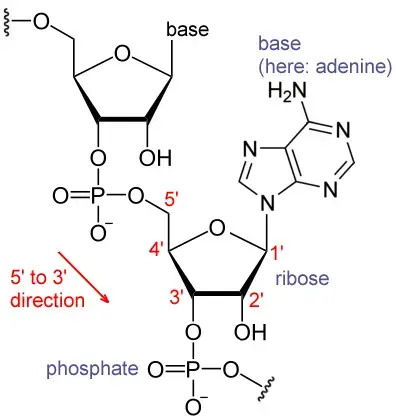
- Polyadenylation is the process of adding a poly(A) tail to an RNA transcript, often messenger RNA (mRNA).
- The poly(A) tail is made up of numerous adenosine monophosphates; in other words, it is an RNA segment with solely adenine nucleotides.
- Polyadenylation is part of the process by which eukaryotes synthesise mature mRNA for translation. In numerous bacteria, the poly(A) tail accelerates mRNA degradation. Consequently, it is a component of the wider process of gene expression.
- The polyadenylation process begins when transcription of a gene concludes. A collection of proteins first cleaves off the 3′-most section of newly synthesised pre-mRNA; these proteins then construct the poly(A) tail at the RNA’s 3′ end.
- These proteins add a poly(A) tail to certain genes at one of several potential locations.
- Similar to alternative splicing, polyadenylation can generate many transcripts from a single gene (alternative polyadenylation).
- Poly(A) tail is essential for nuclear export, translation, and mRNA stability. mRNA is destroyed enzymatically when the tail is sufficiently truncated.
- In a few cell types, however, mRNAs with short poly(A) tails are retained in the cytoplasm for subsequent activation by re-polyadenylation.
- In contrast, bacterial polyadenylation increases the destruction of RNA. Occasionally, this is also the case for eukaryotic noncoding RNAs.
- Both prokaryotes and eukaryotes contain polyadenylated 3′-ends on their mRNA molecules, with the prokaryotic poly(A) tails being shorter and fewer mRNA molecules being polyadenylated.
Poly(A) Polyadenylation Factors
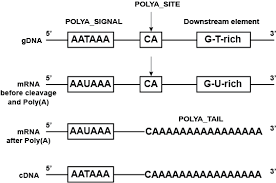
- Cleavage and polyadenylation specificity factor (CPSF) – 360 kDa, which is composed of four distinct polypeptides, first forms an unstable compound with the upstream AAUAAA poly(A) signal.
- Cleavage stimulatory factor (CS1F) – a 200-kDa heterotrimer, interacts with the GU-rich sequence
- Cleavage factor I (CFI) – a 150-kDa heterotrimer
- Cleavage factor II (CFII)
- Poly(A) polymerase (PAP) -binds to the short A tail first supplied by PAP and links cleavage and polyadenylation in a template-independent manner. The substrate is ATP Poly(A)-binding protein II (PABPII).
Mechanism of Polyadenylation
- The processive polyadenylation complex in the nucleus of eukaryotes functions on precursor mRNA and other RNA polymerase II products.
- A multiprotein complex cleaves the 3′-most portion of newly synthesised RNA and polyadenylates the resulting end.
- The enzyme CPSF is responsible for the cleavage, which occurs 10–30 nucleotides downstream of its binding site.
- This location typically contains the polyadenylation signal sequence AAUAAA on the RNA, however variations exist that bind CPSF less strongly.
- Two more proteins, CstF and CFI, give specificity to the RNA-binding process. CstF binds to a GU-rich region downstream of the CPSF binding site.
- CFI recognises a third location on the RNA (a collection of UGUAA sequences in mammals) and can recruit CPSF even in the absence of the AAUAAA sequence.
- The polyadenylation signal, the sequence motif recognised by the RNA cleavage complex, varies among eukaryotic species. The majority of human polyadenylation sites include the sequence AAUAAA, although this pattern is less prevalent in plants and fungi.
- As CstF also attaches to RNA polymerase II, RNA is often cleaved prior to transcription termination.
- Through a process that is poorly understood (as of 2002), it stimulates RNA polymerase II to detach from the transcript.
- The protein CFII is also involved in cleavage, however it is uncertain how. The cleavage location associated with a polyadenylation signal can range between approximately 50 nucleotides.
- Polyadenylation begins upon RNA cleavage, mediated by polyadenylate polymerase.
- Polyadenylate polymerase forms the poly(A) tail by cleaving pyrophosphate from adenosine triphosphate and adding adenosine monophosphate units to the RNA.
- Another protein, PAB2, binds to the new, short poly(A) tail and enhances polyadenylate polymerase’s affinity for RNA.
- When the poly(A) tail reaches around 250 nucleotides in length, the enzyme loses its ability to bind to CPSF and polyadenylation ceases, hence dictating the length of the poly(A) tail.
- CPSF interacts with RNA polymerase II, enabling it to tell the polymerase to stop transcription.
- The end of transcription is signalled when RNA polymerase II encounters a “termination sequence” (5’TTTATT3′ on the DNA template and 5’AAUAAA3′ on the primary transcript).
- The polyadenylation machinery is also physically related to the spliceosome, a complex that eliminates RNA introns.
Poly(A) Polyadenylation in bacteria
- Polyadenylation can also occur in bacterial mRNA.
- Some mRNA in E. coli undergoes polyadenylation, resulting in the creation of a poly A tail of 10 to 40 nucleotides.
- It is triggered by poly A polymerase connected to ribosomes
- Here, the poly A tail serves as a binding site for the nucleases (PNPase and Rnase) that degrade mRNA.
Alternative polyadenylation
- Some protein-coding genes in mRNAs contain multiple polyadenylation sites. Thus, a gene can code for many mRNAs with differing poly A tail positions. This mechanism is known as alternative polyadenylation; it results in the production of several transcripts from a single gene.
- Example: During the growth of B lymphocytes, the synthesis of antibody molecules transitions from membrane-bound to secretory forms.
Cytoplasmic polyadenylation
- In the cytoplasm of certain animal cell types, polyadenylation occurs, specifically in the germ line, during early embryogenesis, and at post-synaptic locations in nerve cells.
- This extends the poly(A) tail of an mRNA with a short poly(A) tail, allowing the mRNA to be translated.
- These truncated poly(A) tails typically consist of fewer than 20 nucleotides and are prolonged to between 80 and 150 nucleotides.
- Even though transcription does not begin until the middle of the 2-cell stage, cytoplasmic polyadenylation of maternal RNAs from the egg cell helps the cell to survive and expand in the early mouse embryo (4-cell stage in human).
- In the brain, cytoplasmic polyadenylation is active during learning and may play a role in long-term potentiation, which is the strengthening of signal transmission in response to nerve impulses and is essential for learning and memory formation.
- Cytoplasmic polyadenylation necessitates the RNA-binding proteins CPSF and CPEB, and can involve additional RNA-binding proteins such as Pumilio.
- The polymerase can be the nuclear polyadenylate polymerase (PAP) or the cytoplasmic polymerase GLD-2, depending on the cell type.
Functions of Poly(A) Polyadenylation
- The poly(A) tail on mRNAs shields the mRNA molecule from enzymatic destruction in the cytoplasm and aids in the termination of transcription.
- mRNA exportation from the nucleus.
- Assist with translation.
What is Deadenylation?
- Most mRNAs in the cytoplasm of eukaryotic somatic cells have poly(A) tails that get shorter over time. mRNAs with shorter poly(A) tails are translated less and broken down faster.
- But it can take hours for an mRNA to be broken down.
- MicroRNAs that match the 3′ untranslated regions of an mRNA can speed up this process of deadenylation and degradation.
- mRNAs with shorter poly(A) tails are not broken down in immature egg cells. Instead, they are stored and cannot be translated.
- After fertilisation, polyadenylation in the cytoplasm of the egg turns on these short-tailed mRNAs.
- Poly(A) ribonuclease (PARN) can attach to the 5′ cap and take nucleotides out of the poly(A) tail in animals.
- How quickly the mRNA breaks down is affected by how easy it is to get to the 5′ cap and poly(A) tail. When the initiation factors 4E and 4G bind to the RNA at the 5′ cap and poly(A) tail, respectively, PARN deadenylates less. This is why translation reduces deadenylation.
- RNA-binding proteins may also be able to change the rate of deadenylation. Also, RNA triple helix structures and RNA motifs like the poly(A) tail 3′ end binding pocket slow down the process of deadenylation and stop the poly(A) tail from being taken off.
- After the poly(A) tail is taken off, the decapping complex takes off the 5′ cap. This causes the RNA to break down.
- Several other proteins, such as the CCR4-Not complex, are also involved in deadenylation in budding yeast and human cells.
References
- Molecular Biology (WCB CELL & MOLECULAR BIOLOGY) by Robert F. Weaver (Author)
- Decroly, E., Ferron, F., Lescar, J. et al. Conventional and unconventional mechanisms for capping viral mRNA. Nat Rev Microbiol 10, 51–65 (2012). https://doi.org/10.1038/nrmicro2675
- https://www.sparknotes.com/biology/molecular/posttranscription/section1/
- https://www.slideshare.net/EmaSushan/5-capping
- https://www.slideshare.net/IndrajaDoradla/rna-processing-238362419