Bacterial transformation is a process wherein bacteria naturally modify their genetic makeup by incorporating foreign genetic material into their own genome. This phenomena has significant ramifications for health care, agriculture, and environmental science and has evolved into a fundamental genetic engineering tool. Scientists can create life-saving medications, alter crops for improved features, and speed up environmental cleaning by inserting desired genes into bacteria. In this article, we examine the workings and uses of bacterial transformation, emphasizing how it has the power to alter many different branches of knowledge.
Definition of Bacterial Transformation – What is Bacterial Transformation?
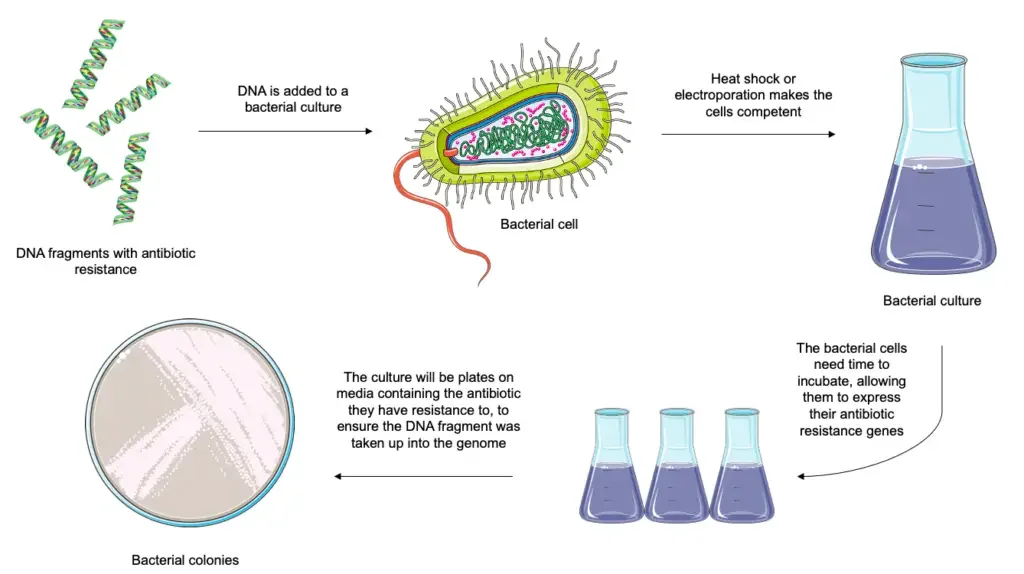
Bacterial transformation is a biological process whereby foreign DNA molecules are taken up and incorporated into the genetic material of the bacterium, changing the genetic makeup of the bacteria. Bacteria can pick up new features or genetic information from their surroundings according to a natural mechanism. This procedure is essential to genetic engineering and has changed a number of academic disciplines by making it possible to introduce particular genes into bacteria for use in environmental research, agriculture, and medicine. Bacterial transformation is a crucial technique for modifying bacterial genomes and researching the expression, function, and inheritance of genes.
Bacterial Transformation Principle
- Bacterial transformation relies on bacteria’s inherent ability to liberate DNA, which is then taken up by another competent bacterium.
- Transformation is dependent upon the competence of the host cell. Competence is a cell’s ability to assimilate naked DNA during the transformation process.
- During the late stationary phase, naturally transformable organisms liberate their DNA through autolysis.
- Several bacteria, including Escherichia coli, can be artificially treated in the laboratory to enhance their transformability through the application of chemicals, such as calcium, a strong electric field (electroporation), or a heat shock.
- Electroporation or thermal shock increases competence by increasing the permeability of the cell wall, allowing the donor DNA to enter the cell.
- Transformants can also be chosen if the transformed DNA contains a selectable marker, such as antimicrobial resistance, or if the DNA encodes for utilization of a growth factor, such as an amino acid.
- In the majority of naturally competent bacteria, liberated DNA binds to the bacteria and is incorporated into chromosomal DNA.
- Occasionally, the free DNA is inserted into a plasmid capable of replicating independently from the chromosome; therefore, the insert does not need to be integrated into the chromosome.
- Some enzymes and antibiotic-resistant markers encoded on the plasmid are expressed in the transformant after transformation.
- In this transformation procedure, donor DNA is initially inserted into the plasmid. The donor DNA-containing plasmid is then introduced into the competent host bacteria.
- Upon completion of the transformation, plasmid-containing bacteria can be detected using a growth medium supplemented with a specific antibiotic.
Before starting transformation
Prepare and allow to set LB agar plates. If pre-poured platters are to be used, they must be heated to 37 degrees Celsius. Incorporate the appropriate antibiotic into the LB agar based on the antibiotic marker present in the plasmid DNA. If blue/white screening for recombinants is necessary, LB agar should contain 1 mM IPTG, 300 g/mL S-Gal or 40 g/mL X-Gal, and 500 g/mL ferric ammonium citrate.
If electrocompetent cells are used, the electroporation chamber should be placed on cold. Bring the water temperature to 37 degrees Celsius. Bring the sterile SOC medium to room temperature (or 20 to 25 degrees Celsius in a water immersion).
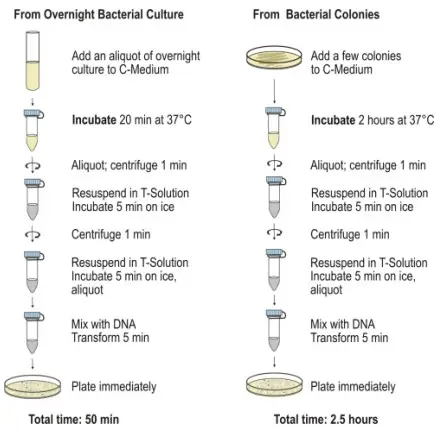
Protocol For Transformation Using Chemically Competent Cells
Materials required
| Reagents | Equipment |
|---|---|
| SOC mediu | Shaker incubator (37 °C) |
| LB agar EZMix™ | Cabinet incubator (37 °C) |
| Appropriate selection antibiotic | Heated water bath (37 °C) |
| IPTG (Isopropyl-β-D-thiogalactoside) | 15 mL polypropylene culture tubes (sterile) |
| X-gal | Culture dishes |
| S-Gal™ | Lazy-L sterile spreaders |
| Ferric Ammonium citrate |
Standard Transformation Protocol
- Transfer the necessary number of tubes from the freezer at -70°C to damp ice. Include an extra tube for control DNA, if desired
- Permit the cells to defrost for 5 minutes. Tap the passages repeatedly to obtain a uniform suspension.
- For control: In one tube, add 1 µL (10 ng) of pUC19 control DNA. Mix by tapping gently and placing on ice
- 1 ng to 50 ng of purified plasmid DNA should be added directly to the cells in the remaining containers. Mix by tapping gently and placing on ice
- Refrigerate the cells for 30 minutes.
- 45 seconds after transferring the cells to a 37 °C water immersion
- Transfer the cells for 2 minutes to cold
- Incorporate SOC medium into each tube. Transfer the cells to sterile polypropylene tubes and loosen the lids to promote culture aeration.
- The cells are incubated on a vibratory incubator (225-250 rpm) at 37°C for one hour.
- Spread 10-100 µL of each transformed cell suspension using a sterile spreader on LB agar plates containing a selection antibiotic.
- Incubate plates overnight at 37°C
- Choose the desired colony(ies) and culture.
- Each culture’s plasmid DNA should be isolated.
- Cultivate the desired clones
- Utilizing restriction enzymes, separate the plasmid DNA using gel electrophoresis.
Protocol For Transformation Using Electrocompetent Cells
Materials required
| Reagents | Equipment |
|---|---|
| SOC medium | Shaker incubator (37 °C) |
| LB agar EZMix™ | Cabinet incubator (37 °C) |
| Appropriate selection antibiotic | Heated water bath (37 °C) |
| IPTG (Isopropyl-β-D-thiogalactoside) | 15 mL polypropylene culture tubes (sterile) |
| X-gal | Culture dishes |
| S-Gal™ | Lazy-L sterile spreaders |
| Ferric Ammonium citrate | Culture dishes |
Standard Transformation Protocol
- Transfer the necessary number of tubes from the freezer at -70°C to damp ice. If desired, include an additional vial for control DNA.
- Permit the cells to defrost for 5 minutes. Tap the tubes gently multiple times to achieve a uniform suspension.
- Transfer SOC medium to individual culture tubes for each transformation reaction and store at room temperature.(The volume of SOC medium is dependent on the number of cells added in the subsequent phase. The final volume with competent cells and SOC medium should be one thousand microliters (L).
- One cuvette and microcentrifuge tube should be placed on cold for each transformation reaction.
- Transfer the competent cells to microcentrifuge containers that have been chilled. Utilize 40 µL of cells from an 80 µL vial and 50 µL of cells from a 100 L vial.
- For control: One vial should contain µ1 L of 1 to 5 dilutions of pUC19 control DNA.
- Prepare a dilution of 1 to 5 folds of DNA or ligation mixture in TE buffer. Combine in microfuge containers.
- The DNA and cell mixture is pipetted into a chilled 1 mm cuvette.
- Treat the cells by setting the electroporator to a field strength of 25 kV/cm for 6 milliseconds.
- Remove cells from cuvettes and add them to SOC medium-filled tubes.
- At 37°C, incubate the cells on a vibratory incubator (225-250 rpm) for one hour.
- Spread 10-100 µL of each transformed cell suspension using a sterile spreader on LB agar plates containing a selection antibiotic.
- Incubate plates overnight at 37° C.
- Select the appropriate colony and culture.
- Isolate the DNA from plasmids in each culture.
- Separate the plasmid DNA using gel electrophoresis after restriction enzyme digestion.
- Cultivate the desired clones.
Factors Affect Transformation Efficiency
Several factors can influence the efficacy of genetic transformation, or the rate at which foreign DNA is successfully integrated into the genome of the host organism. These elements include:
- Competent cells: Competent cells refer to the capacity of bacterial cells to absorb exogenous DNA. Not all bacterial strains are inherently competent, so it is often necessary to create competent cells using laboratory techniques, such as chemical treatment or electroporation. The level of competence of cells can vary, with a higher level of competence generally resulting in greater transformation efficacy.
- DNA quality: The quality and integrity of the introduced DNA play a vital role in transformation efficiency. Pure, undamaged, and highly concentrated DNA has a greater chance of effectively integrating into the host genome.
- DNA fragment size: DNA fragment size can impact transformation efficiency. In general, smaller DNA fragments have a greater transformation efficacy than larger fragments. It may be more challenging for larger fragments to traverse the bacterial cell membrane and integrate into the genome.
- Concentration of DNA: The DNA concentration used for transformation can have an effect on its efficacy. Increasing the DNA concentration can increase the likelihood of successful incorporation into the host genome, up to a certain threshold beyond which it may become inhibitive.
- Technique of transformation: The technique used to introduce DNA into host cells can have a significant impact on transformation efficiency. Chemical transformation, electroporation, and biolistic (gene gun) methods are typical. Each method has its own benefits and drawbacks, and the choice of method is contingent on the organism and DNA being used.
- Growth conditions: The growth conditions of the host cells before and during transformation can have an effect on their efficacy. Temperature, the composition of the medium, and the growth phase can influence the competence of the cells and their capacity to take up DNA.
- Selection and screening: The application of selective pressure, such as antibiotics or markers, can assist in distinguishing transformed cells from nontransformed cells. The accuracy and efficiency of identifying effectively transformed cells are affected by the selection of suitable selection markers and screening techniques.
How is Transformation Efficiency Calculated ?
Transformation efficiency is typically calculated by determining the number of successful transformants (cells that have taken up and incorporated the foreign DNA) per unit of input DNA or per unit of competent cells used in the transformation process. The calculation involves the following steps:
- Determine the number of viable transformants: After the transformation process, the transformed cells are usually plated onto selective agar plates containing antibiotics or other selection markers to differentiate them from non-transformed cells. After an appropriate incubation period, visible colonies representing viable transformants will appear on the plates.
- Count the number of transformant colonies: Count the number of individual transformant colonies on the selective agar plates. To ensure accuracy, it is often necessary to dilute the transformed cell suspension and plate appropriate dilutions to obtain plates with a countable number of colonies (typically between 30 and 300 colonies).
- Calculate the transformation efficiency: The transformation efficiency is typically expressed as the number of transformants per microgram (μg) of input DNA or per nanogram (ng) of input DNA. The formula for calculating transformation efficiency is: Transformation efficiency = (Number of transformants / Amount of input DNA) × Dilution factor. Alternatively, if transformation efficiency is calculated per unit of competent cells, the formula becomes: Transformation efficiency = (Number of transformants / Volume of competent cells) × Dilution factor. The dilution factor accounts for the dilution used for plating an appropriate number of transformant colonies.
It’s important to note that transformation efficiency can vary depending on the specific experimental conditions, such as the strain of bacteria, the type and quality of DNA, the transformation method used, and other factors discussed earlier. Calculating and comparing transformation efficiencies can help optimize transformation protocols and assess the effectiveness of different experimental variables.
Important tips for optimization of transformation process:
- Confirm upon receipt that the cells are still frozen and that dry ice is present.
- Use a high-quality DNA sample devoid of phenol, ethanol, proteins, ions, and detergents for optimal transformation efficiency. A high salt concentration in the DNA of electrocompetent cells will cause arcing at high voltage, which may damage the sample and the apparatus.
- For transformation, DNA in ligation reactions containing high-quality reagents may be used. There is no need to deactivate the DNA ligase.
- Maintain the competent cells on ice at all times; avoid accidental warming.
- Tap the tubes lightly to obtain a uniform cell suspension. Do not combine with a vortex or pipette.
- For optimum colony growth, heat LB agar plates to 37 °C.
- Configuration of the electroporator for transformation of electrocompetent cells:
- BTX Model ECM 630: HV mode, 2.5 kV, 25 µF, 100 Ω, 1 mm cuvette
- BioRad Gene Pulser: 2.5 kV, 25 µF, 100 Ω, 1 mm cuvette
References
- Banerjee, Ratul. (2022). What is genetic transformation as a gene delivery mechanism and how to calculate transformation efficiency?.
- Ren, Jun & Karna, Sandeep & Lee, Hyang-Mi & Yoo, Seung & Na, Dokyun. (2019). Artificial transformation methodologies for improving the efficiency of plasmid DNA transformation and simplifying its use. Applied Microbiology and Biotechnology. 103. 1-11. 10.1007/s00253-019-10173-x.
- Froger A, Hall JE. Transformation of plasmid DNA into E. coli using the heat shock method. J Vis Exp. 2007;(6):253. doi: 10.3791/253. Epub 2007 Aug 1. PMID: 18997900; PMCID: PMC2557105.
- https://goldbio.com/articles/article/Bacterial-Transformation-Deep-Dive
- https://www.phdnest.com/bacterial-transformation/
- https://www.addgene.org/protocols/bacterial-transformation/
- https://www.thermofisher.com/in/en/home/life-science/cloning/cloning-learning-center/invitrogen-school-of-molecular-biology/molecular-cloning/transformation/competent-cell-basics.html
- https://www.sigmaaldrich.com/IN/en/technical-documents/technical-article/genomics/cloning-and-expression/competent-cells
- https://www.sigmaaldrich.com/IN/en/technical-documents/technical-article/genomics/cloning-and-expression/competent-cells
- https://www.assaygenie.com/blog/guide-to-bacterial-transformation-the-science-of-genetic-manipulation
- https://www.assaygenie.com/blog/guide-to-bacterial-transformation-the-science-of-genetic-manipulation
- https://www.sigmaaldrich.com/IN/en/technical-documents/technical-article/genomics/cloning-and-expression/competent-cells
- https://international.neb.com/protocols/2012/05/21/transformation-protocol
- https://www.protocols.io/view/bacterial-transformation-protocol-bp2l6nb9kgqe/v1
- https://www.protocols.io/view/bacterial-transformation-protocol-bp2l6nb9kgqe/v1
- https://scigine.com/blog/bacterial-transformation-protocol-competent-cells/
- https://www.genscript.com/transformation-troubleshooting-guide.html
- https://www.teachingexpertise.com/classroom-ideas/transforming-activity/
- https://www.semanticscholar.org/paper/Bacterial-Transformation%3A-What-Why-How-and-When-Das-Raythata/cc2a9a481f4f336b0e4cf14da9549acf53b37ebb
- https://www.semanticscholar.org/paper/Bacterial-Transformation%3A-What-Why-How-and-When-Das-Raythata/cc2a9a481f4f336b0e4cf14da9549acf53b37ebb
- https://assets.fishersci.com/TFS-Assets/LSG/manuals/MAN0012725_TransformAid_Bacterial_Transformation_UG.pdf
- https://biology-forums.com/index.php?action=gallery;sa=view;id=97
- https://www.busyinag.com/2021/09/mechanism-of-transformation-in-bacteria.html
- https://www.sciencelearn.org.nz/resources/2032-bacterial-transformation
- http://ncdnaday.org/2020/11/technique-tuesday-bacterial-transformation/
- https://step1.medbullets.com/microbiology/104019/bacterial-genetics