What is chitin?
- Chitin is a big structural polysaccharide composed of glucose chains that have been changed. The exoskeletons of insects, the cell walls of fungi, and certain rigid structures of invertebrates and fish include chitin.
- Chitin ranks second in terms of abundance, behind only cellulose. Around one billion tons of chitin are produced annually by species in the biosphere.
- This incredibly adaptable molecule can build solid structures on its own, as in the case of insect wings, or it can interact with other things, such as calcium carbonate, to produce even stronger compounds, as in the case of a clam’s shell.
- Like cellulose, no vertebrate species can independently digest chitin. Insect-eating animals typically have symbiotic bacteria and protozoa that can degrade chitin’s fibrous structure into its component glucose molecules.
- Yet, because chitin is a biodegradable molecule that degrades over time, it is employed in a variety of industrial applications, including surgical thread and dye and glue binders.
Structure of chitin
- N-acetyl-D-glucosamine is poly-(1→4)-β-linked to chitin (42). The sugar units in its structure are rotated by 180°, and each pair creates the disaccharide N,N′-diacetylchitobiose [(GlcNAc)2].
- The individual polymer chains are helices in which each sugar unit is inverted relative to its neighbors. As a result of the 03-H→05 and 06-H→07 hydrogen bonds connecting the stiff ribbons, this structure is highly stable.
- Moreover, chitin has three distinct crystalline allomorphs: the α-, β- and γ-forms. The orientation of the microfibrils differs between these two types. The most prevalent kind of chitin is -chitin. Its unit cell consists of two N,N′-diacetylchitobiose units arranged antiparallel to one another.
- Consequently, adjacent polymer chains are linked together by 06-H→06 hydrogen bonds, and the chains are maintained in sheets by 07→H-N hydrogen bonds. This results in a statistical mixture of -CH2OH orientations, with half of the oxygen atoms on each residue capable of forming intermolecular hydrogen bonds.
- Half of the amide groups function as acceptors for C=O→H-N intramolecular hydrogen bonding. The formation of these intermolecular hydrogen bonds results in a structure that is highly stable. If the polymer chains are allowed to solidify, they will self-assemble into microfibrils.
- β-chitin is a less common type of chitin in which the unit cell is an N,N′-diacetylchitobiose unit, resulting in a polymer stabilized as a stiff ribbon by 03→05 intramolecular H-bonds, the same as -chitin.
- The sheets of chains in this structure are held together by C=O→H-N H-bonds between the amide groups and by the -CH2OH side chains, which leads to the formation of intersheet H-bonds to the carbonyl oxygens on the neighboring chains (06-H→07).
- This results in a structure of parallel poly-N-acetylglucoseamine chains with no H-bonds between sheets. The parallel organization of polymer chains in -chitin allows for greater flexibility than the antiparallel arrangement of polymer chains in β-chitin, yet the resulting polymer retains enormous strength.
- The third allomorph with mixed parallel and antiparallel orientations is -chitin. It has reportedly been found in mushroom. With the exception of the β-chitin present in diatoms, chitin is always cross-linked to other structural components. In fungal cell walls, chitin is covalently attached to glucans, either directly, as in Candida albicans, or via peptide bridges.
- Furthermore, in insects and other invertebrates, chitin is always covalently and noncovalently bound to certain proteins. This relationship indicates that the observed orderly structures are produced. There are also different levels of mineralization, including calcification and sclerotization, which involve interactions with phenolic and lipid molecules.
- In animals such as fungi and invertebrates, various degrees of deacetylation have been documented, creating a continuum of structure between chitin (completely acetylated) and chitosan (completely deacetylated).
- Although either acids or alkalis can be used to deacetylate chitin, due to the susceptibility of glycosidic linkages to acid, which would break the chain, alkali deacetylation is more commonly utilized.
- Chitin can be N-deacetylated using either heterogeneous or homogeneous reaction mixtures. There is no precise differentiation between chitin and chitosan with varying degrees of deactylation. Exceptions apart, natural chitin exists in association with other structural polymers such as proteins or glucans, which frequently comprise more than 50 percent of the mass in chitin-containing tissue.
- Chitin can be N-deacetylated to the point of solubility in diluted acetic and formic acids. In chitin, acetylated units predominate and the degree of acetylation is often 0.90, whereas chitosan is a wholly or partially N-deacetylated derivative with a degree of deacetylation that is typically greater than 0.65.
- Numerous analytical techniques, including IR spectroscopy, pyrolysis gas chromatography, gel permeation chromatography and UV-vis spectrophotometry, 1H NMR spectroscopy, 13C solid state NMR, thermal analysis, various titration schemes, acid hydrolysis, HPLC, separation spectrometry methods, and more recently near-infrared spectroscopy, have been used to determine the degree of deacetylation.
What are Chitinases?
- Chitinases (EC 3.2.1.14, chitodextrinase, 1,4-β-poly-N-acetylglucosaminidase, poly-β-glucosaminidase, β-1,4-poly-N-acetyl glucosamidinase, poly [1,4-(N-acetyl-β-D-glucosaminide)] glycanohydrolase, (1→4) -2-acetamido-2-deoxy-β-D-glucan glycanohydrolase; systematic name (1→4) -2-acetamido-2-deoxy-β-D-glucan glycanohydrolase) hydrolytic enzymes that degrade chitin’s glycosidic linkages. They trigger the subsequent reaction:
- Endohydrolysis of N-acetyl-β—D-glucosaminide (1→4)-β—linkages in chitin and chitodextrins occurs at random.
- Although chitin is a component of the cell walls of fungus and the exoskeletons of certain animals (including mollusks and arthropods), chitinases are typically found in organisms that either need to reconfigure their own chitin or breakdown and digest the chitin of fungi or animals.
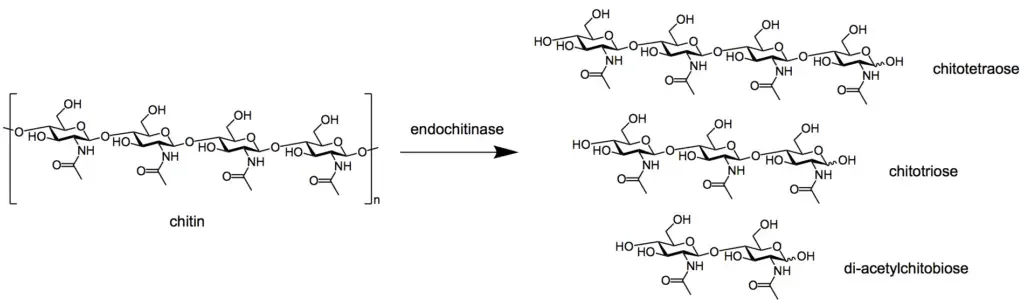
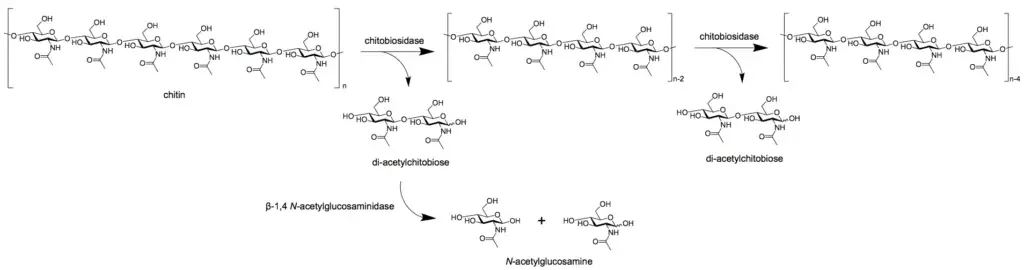
- Endochitinases (EC 3.2.1.14) randomly cleave chitin at interior locations of the chitin microfibril, producing soluble, low-molecular-weight multimer products. Di-acetylchitobiose, chitotriose, and chitotetraose are the multimer products, with the dimer being the most abundant.
- Moreover, exochitinases have been separated into two subcategories:
- Chitobiosidases (EC 3.2.1.29) work on the non-reducing end of the chitin microfibril, releasing di-acetylchitobiose from the chitin chain one molecule at a time. In this process, there is no release of monosaccharides or oligosaccharides .
- β-1,4- N-acetylglucosaminidases (EC 3.2.1.30) break down the multimer products, such as di-acetylchitobiose, chitotriose, and chitotetraose, into N-acetylglucosamine monomers (GlcNAc).
- Chitinases were also classified based on their amino acid sequences, as this would be more helpful in understanding the evolutionary relationships between these enzymes. Therefore, chitinases were grouped into three families: 18, 19, and 20.
- Families 18 and 19 contain endochitinases from a variety of organisms, including viruses, bacteria, fungi, insects, and plants. Nevertheless, family 19 consists primarily of chitinases from plants. N-acetylglucosaminidase and a related enzyme, N-acetylhexosaminidase, are members of Family 20.
- Moreover, since the gene sequences of the chitinases were known, they were divided into six groups based on their sequences. N-terminal sequence, enzyme location, isoelectric pH, signal peptide, and inducers were characteristics that characterized the classes of chitinases.
- Class I chitinases were characterized by a cysteine-rich N-terminus, a leucine- or valine-rich signal peptide, and vacuolar location. Next, based on their acidic or basic nature, Class I chitinases were further split into Class Ia and Class Ib, respectively. Class 1 chitinases were found to contain only plant chitinases and predominantly endochitinases.
- Class II chitinases lacked the cysteine-rich N-terminus but shared the same sequence as Class I enzymes. Class II chitinases were predominantly exochitinases and were discovered in plants, fungus, and bacteria.
- Class III chitinases did not have sequence similarities with Class I or Class II chitinases.
- Class IV chitinases shared similar traits, particularly immunological qualities, with Class I chitinases; however, Class IV chitinases were considerably smaller in size than Class I chitinases.
- Chitinases of Classes V and VI are poorly described. However, one example of a Class V chitinase displayed two chitin binding domains in tandem, and based on the gene sequence, the cysteine-rich N-terminal appeared to have been lost during evolution, most likely as a result of decreased selective pressure causing the catalytic domain to lose its function.
Factors affecting chitin degradation
Chitin degradation can be influenced by several factors, including:
- Enzymatic activity: Chitinases, enzymes that break down chitin, can be produced by various microorganisms, fungi, and animals. The presence and activity of these enzymes can greatly affect chitin degradation.
- pH: Chitin degradation can be affected by the pH of the environment. Chitinases generally have an optimum pH range, and changes in pH can affect their activity.
- Temperature: Like pH, temperature can also affect chitinase activity. Most chitinases have an optimum temperature range, and changes in temperature can affect their efficiency.
- Moisture content: Chitin is insoluble in water, so moisture content can play a role in chitin degradation. Moisture can help facilitate enzymatic activity and improve the accessibility of chitin to degrading agents.
- Chemical treatment: Chemical treatments, such as acid or alkali hydrolysis, can be used to degrade chitin. However, these treatments can also affect the structure and properties of chitin, making it less useful for certain applications.
- Physical treatment: Physical treatments, such as grinding or milling, can also break down chitin. However, these treatments can also damage the structure of chitin, making it less useful for certain applications.
- Added glucose: Glucose is a simple sugar that can provide a source of energy for microorganisms that produce chitinase. Adding glucose to a chitin-containing environment can increase the activity of chitinase and thus promote chitin degradation.
- Aeration: Aerobic conditions, where oxygen is present, can promote chitin degradation. Oxygen is necessary for the growth and activity of aerobic microorganisms that produce chitinase. Anaerobic conditions, on the other hand, can limit chitin degradation.
- Organic matter: The presence of organic matter, such as decaying plant or animal material, can stimulate the growth of microorganisms that produce chitinase. Organic matter can also provide a source of nutrients and energy for chitinase-producing microorganisms, thus promoting chitin degradation.
Microorganisms Involved in Chitin Degradation
Chitin, a complex polysaccharide found in the cell walls of fungi, insects, and other invertebrates, is an important source of carbon and nitrogen for many microorganisms. Several types of microorganisms are involved in chitin degradation, including chitinolytic bacteria, chitinolytic fungi, slime molds, protozoa, and algae.
- Chitinolytic bacteria: Chitinolytic bacteria are capable of producing chitinases, enzymes that break down chitin into smaller, more easily metabolized molecules. These bacteria are found in many environments, including soil, water, and animal guts. Examples of chitinolytic bacteria include Bacillus, Pseudomonas, and Serratia.
- Chitinolytic fungi: Chitinolytic fungi are also important players in chitin degradation. These fungi produce chitinases and other enzymes that break down chitin, and they are found in many environments, including soil, water, and animal guts. Examples of chitinolytic fungi include Aspergillus, Penicillium, and Trichoderma.
- Slime molds: Slime molds are another group of organisms that can break down chitin. These organisms are typically found in soil and leaf litter, and they are capable of producing enzymes that break down chitin.
- Protozoa: Protozoa, including amoebae and ciliates, are also capable of breaking down chitin. These organisms are found in many environments, including soil and aquatic environments, and they are important for recycling nutrients.
- Algae: Finally, some algae are also capable of breaking down chitin. These organisms are typically found in aquatic environments and are important for nutrient cycling.
In summary, chitin degradation is carried out by a variety of microorganisms, including chitinolytic bacteria, chitinolytic fungi, slime molds, protozoa, and algae. These organisms play an important role in nutrient cycling and the breakdown of organic matter in many ecosystems.
Enzymes involved in the degradation of chitin
Chitin, a complex polysaccharide found in the cell walls of fungi, insects, and other invertebrates, is degraded by a variety of enzymes produced by different microorganisms. The main enzymes involved in chitin degradation are chitinases and chitinase-like proteins, as well as lytic polysaccharide monooxygenases (LPMOs).
- Chitinases are enzymes that specifically hydrolyze the beta-1,4 glycosidic bonds in chitin, breaking it down into smaller, more easily metabolized molecules. These enzymes are produced by a wide range of organisms, including bacteria, fungi, plants, and animals. Chitinases are classified into different families based on their amino acid sequences, and they vary in their substrate specificities and other properties.
- Chitinase-like proteins (CLPs) are a group of proteins that are structurally similar to chitinases but do not have chitinase activity. These proteins are thought to play a role in the regulation of chitinase activity, as well as in other processes.
- Lytic polysaccharide monooxygenases (LPMOs) are another group of enzymes that are involved in chitin degradation. These enzymes are capable of oxidizing the glycosidic bonds in chitin, making them more susceptible to hydrolysis by chitinases and other enzymes. LPMOs are produced by a variety of organisms, including bacteria, fungi, and animals.
- Other enzymes that may be involved in chitin degradation include proteases, which break down chitin-binding proteins and other proteins associated with chitin, and esterases, which cleave the ester bonds between N-acetylglucosamine residues in chitin.
In summary, chitin is degraded by a variety of enzymes produced by different microorganisms, including chitinases, chitinase-like proteins, lytic polysaccharide monooxygenases, proteases, and esterases. These enzymes play an important role in nutrient cycling and the breakdown of organic matter in many ecosystems.
Simple Steps of chitin degradation
Chitin degradation typically occurs in a two-step process: depolymerization and deacetylation.
- Step 1: Depolymerization: During depolymerization, chitinase enzymes break down the long chains of chitin molecules into smaller fragments. This process involves the cleavage of the beta-1,4 glycosidic bonds that link the N-acetylglucosamine (GlcNAc) monomers in the chitin chains. The resulting products are shorter chitin oligomers, such as chitotriose, chitobiose, and monomeric GlcNAc.
- Step 2: Deacetylation: Deacetylation involves the removal of the acetyl groups from the GlcNAc monomers, resulting in the production of chitosan. This process is typically carried out by the action of chitinases and chitosanases, as well as by chemical and physical methods. The degree of deacetylation can vary depending on the conditions used, and it can affect the properties of the resulting chitosan product.
Overall, chitin degradation is a complex process that can be influenced by many factors, and the specific steps involved can vary depending on the conditions and methods used. However, depolymerization and deacetylation are two key steps that are typically involved in the breakdown of chitin into its constituent components.
Mechanisms of microbial degradation of chitin
Microbial degradation of chitin can occur through one of two mechanisms: chitinoclastic or deacetylation.
- Chitinoclastic Mechanism: The chitinoclastic mechanism involves the enzymatic breakdown of chitin by chitinase enzymes, which cleave the beta-1,4 glycosidic bonds between the N-acetylglucosamine (GlcNAc) monomers in the chitin molecule. This process produces shorter chitin oligomers, such as chitotriose and chitobiose, as well as monomeric GlcNAc. These shorter chitin fragments can then be further degraded by other chitinases or utilized as a source of carbon and nitrogen by chitinolytic microorganisms.
- Deacetylation Mechanism: The deacetylation mechanism involves the removal of acetyl groups from GlcNAc monomers in chitin molecules. This process is typically carried out by the action of chitinases and chitosanases, as well as chemical and physical methods. The removal of acetyl groups results in the production of chitosan, which has different properties than chitin and can be further degraded by chitosanases or utilized as a source of carbon and nitrogen by chitinolytic microorganisms.
Both mechanisms can occur simultaneously or sequentially, depending on the specific microorganisms and environmental conditions involved in chitin degradation. Overall, microbial chitin degradation is a complex process that involves multiple enzymes and pathways, and its efficiency can be influenced by many factors, including the presence of co-substrates, environmental conditions, and the activity of other microorganisms in the ecosystem.
FAQ
What is chitin, and why is it important?
Chitin is a polysaccharide that is found in the exoskeletons of arthropods, as well as in the cell walls of fungi and some algae. It is the second most abundant biopolymer on earth after cellulose, and it plays an important role in the ecology of many ecosystems.
How do microorganisms degrade chitin?
Microorganisms can degrade chitin through two main mechanisms: chitinoclastic and deacetylation. Chitinases are the key enzymes involved in chitinoclastic degradation, while chitosanases and other enzymes are involved in deacetylation.
What are some examples of microorganisms that can degrade chitin?
Many microorganisms can degrade chitin, including bacteria, fungi, and some protozoa. Some examples of chitin-degrading microorganisms include Vibrio cholerae, Serratia marcescens, and Trichoderma reesei.
What are the products of chitin degradation?
The products of chitin degradation can vary depending on the specific microorganisms and conditions involved. However, common products include chitin oligomers, chitosan, and monomeric N-acetylglucosamine.
What is the role of chitin-degrading microorganisms in the ecosystem?
Chitin-degrading microorganisms play an important role in the recycling of organic matter in the ecosystem. They break down chitin-containing waste products, such as exoskeletons and fungal cell walls, into simpler compounds that can be utilized by other organisms.
Can chitin-degrading microorganisms be used for biotechnological applications?
Yes, chitin-degrading microorganisms are of interest for various biotechnological applications, such as the production of chitin oligomers, chitosan, and other value-added compounds.
What factors can affect chitin degradation?
Many factors can influence chitin degradation, including pH, temperature, nutrient availability, oxygen levels, and the activity of other microorganisms in the ecosystem.
How is chitin degradation studied in the laboratory?
Chitin degradation can be studied in the laboratory using various techniques, such as enzyme assays, microbial culture techniques, and molecular biology methods.
What are some challenges associated with chitin degradation?
Chitin degradation can be challenging due to the complexity of the chitin molecule, the presence of other polymers in the ecosystem, and the variability of chitin-degrading microorganisms.
What is the potential impact of chitin degradation on global carbon and nitrogen cycling?
Chitin degradation plays an important role in global carbon and nitrogen cycling, as it contributes to the recycling of organic matter in the ecosystem. However, the exact impact of chitin degradation on these processes is still being studied.
References
- Gooday, G.W. Physiology of microbial degradation of chitin and chitosan. Biodegradation 1, 177–190 (1990). https://doi.org/10.1007/BF00058835
- Elieh-Ali-Komi D, Hamblin MR. Chitin and Chitosan: Production and Application of Versatile Biomedical Nanomaterials. Int J Adv Res (Indore). 2016 Mar;4(3):411-427. Epub 2016 Mar 1. PMID: 27819009; PMCID: PMC5094803.
- Beier, S., & Bertilsson, S. (2013). Bacterial chitin degradation—mechanisms and ecophysiological strategies. Frontiers in Microbiology, 4. doi:10.3389/fmicb.2013.00149
- Beier S, Bertilsson S. Bacterial chitin degradation-mechanisms and ecophysiological strategies. Front Microbiol. 2013 Jun 14;4:149. doi: 10.3389/fmicb.2013.00149. PMID: 23785358; PMCID: PMC3682446.
- https://biologydictionary.net/chitin/