What is SDS-PAGE?
- SDS–PAGE refers as a Sodium Dodecyl Sulfate–PolyAcrylamide Gel Electrophoresis, it is widely used technique for separation of proteins according to their molecular weight.
- The technique is based on principle that proteins are separated in electric field by their charge-to-mass ratio, but here, because of SDS, mostly separation depend on molecular size only.
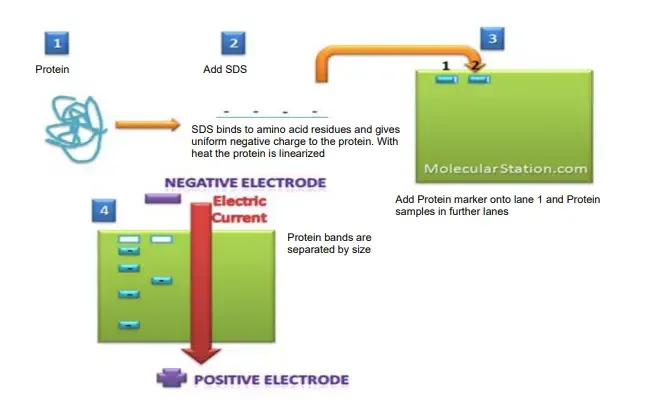
- In this method the detergent SDS (C₁₂H₂₅SO₄Na) binds with proteins, giving them uniform negative charge, which masks their native charge properties.
- As result, movement of proteins in gel matrix is determined mainly by the length of polypeptide chain, not by their shape or charge.
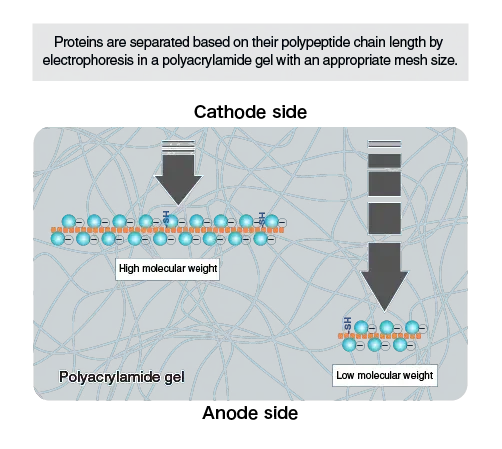
- Usually the gel is prepared with polyacrylamide, which act like a molecular sieve, smaller proteins move faster / farther than larger ones.
- The gel concentration may be adjusted according to size range of proteins to be separated, e.g. higher % gels for small proteins, lower % for large ones.
- Before loading, protein samples are mixed with SDS and often with β-mercaptoethanol or DTT, which help to break disulfide bonds and denature the proteins into linear form.
- During electrophoresis, an electric current is applied, and negatively charged protein–SDS complexes migrate toward the positive electrode (anode).
- After run, the gel is stained with dyes like Coomassie Brilliant Blue or Silver stain, to visualize separated protein bands.
- The distance migrated by each band is inversely related with log of their molecular weight, and unknown proteins can be estimated by comparing with known molecular weight markers.
- This technique has been used for analytical / preparative purpose, protein purity checking, molecular weight estimation, or verifying expression of recombinant proteins etc.
- Some minor variations of SDS–PAGE exist like 2D–PAGE, where proteins are separated first by isoelectric focusing, then by SDS–PAGE, giving higher resolution.
- It’s important that buffer system, pH and running conditions are maintained carefully, else poor resolution or smeared bands may occur.
- Although powerful, it does not give information about protein function or native conformation, because proteins are completely denatured by SDS.
- So, SDS–PAGE mainly used for analytical profiling rather than functional study of protein molecules, and it still remain one of the most sturdy and hardy methods used in biochemical labs worldwide.
Definition of SDS-PAGE
SDS-PAGE is a technique used to separate proteins based on their molecular weight by using a gel matrix and the denaturing agent sodium dodecyl sulfate (SDS).
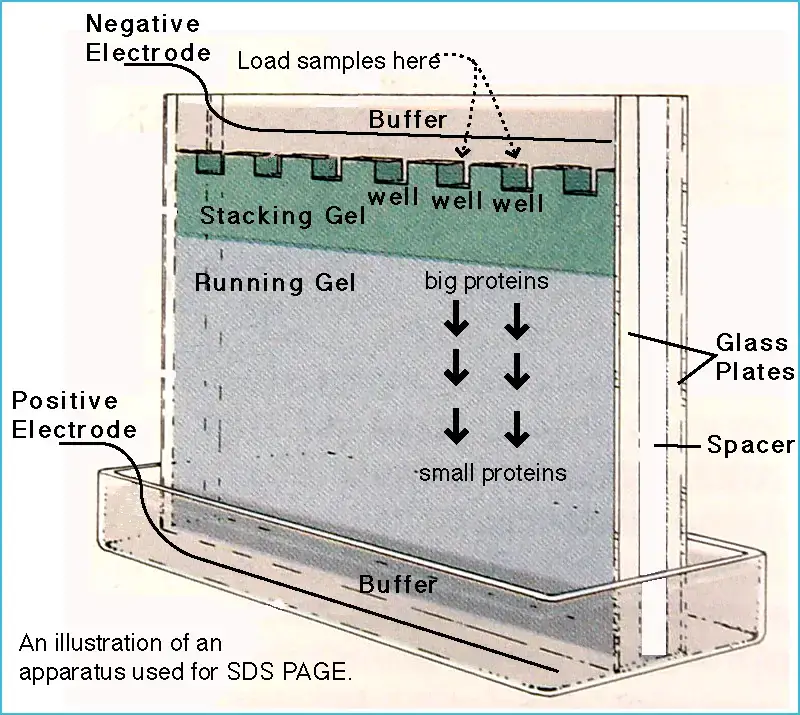
Features
Uniform Negative Charge – In SDS–PAGE, the detergent SDS (sodium dodecyl sulfate) binds to proteins giving them almost same negative charge density, so migration depend mainly by molecular size.
Denaturation of Proteins happen by SDS and sometimes by heating, which unfold the protein structure to linear chain form for equal comparison.
Polyacrylamide Gel Matrix act as molecular sieve; smaller proteins travel faster, while large ones move slower by the pores of gel.
High Resolution Separation is obtained, it allow to differentiate proteins with small difference in molecular mass, sometimes as little as few kDa.
The gel concentration can be adjusted (5–20%) depending upon size of target proteins, this flexibility make the method adaptable.
Tracking Dye like bromophenol blue is usually used to monitor front movement during electrophoresis, though it not affect protein migration.
Sample Buffer usually contains SDS, glycerol, Tris-HCl, bromophenol blue, and β-mercaptoethanol / DTT, all together for denaturation and loading purpose.
Quantitative Estimation of molecular weight can be done by comparing migration distance with known protein markers on same gel.
The technique is reproducible and sturdy, commonly used for routine protein checking in biochemistry / molecular biology labs.
After electrophoresis, staining with dyes (like Coomassie Brilliant Blue or silver stain) used for band visualization, and de-staining done for clarity.
Dual Gel System is often employed – stacking gel (low % acrylamide) and resolving gel (high % acrylamide) to improve band sharpness.
In SDS–PAGE, the mobility of proteins depend logarithmically on molecular mass, that allow linear plot of log(MW) vs migration distance.
Equipment setup involve gel casting plates, electrophoresis tank, buffer system, and power supply, yet they are easy to handle.
pH Stability is maintained by buffer (usually Tris-glycine system), ensuring constant charge during run though sometimes pH gradient may develop.
It’s mainly used for analysis purpose, like checking protein purity, estimation of subunit composition, and comparing expression pattern etc.
SDS–PAGE not suitable for determining native protein structure or enzymatic activity, since SDS cause complete denaturation of tertiary structure.
Despite its simplicity and age, this method still prevail as one of the most widely adopted tools in protein biochemistry, because it’s reliable and cheap to perform.
Objective
- To Separate Proteins according to their molecular weight, by removing the effect of native charge and shape through binding with SDS detergent.
- To Determine Molecular Weight of unknown proteins by comparing their migration distance with known molecular weight standards / markers.
- To Check Purity of protein sample after purification process, since presence of extra bands show contamination or degradation products.
- To Analyze Subunit Composition of multimeric proteins by denaturing them into individual polypeptide chains and separating those by gel.
- To Monitor Protein Expression during recombinant protein production or gene expression studies, by comparing the intensity of target band.
- To Evaluate Efficiency of purification steps like precipitation, chromatography etc., by comparing band pattern before and after each step.
- To Confirm Presence or Absence of specific protein in biological sample by band position and size when compared to control or marker lane.
- To Study Post–Translational Modifications indirectly, since modified proteins (like phosphorylated or glycosylated) show slight shift in mobility.
- To Prepare Protein Samples for further analysis like Western Blotting, where SDS–PAGE act as first step for protein transfer to membrane.
- To Estimate Relative Quantity of proteins by comparing staining intensity between bands within same gel (semi–quantitative purpose).
- To Characterize Protein Mixtures from tissues, cells, or organelles and understand their profile or expression pattern.
- To Provide Visual Representation of protein molecular diversity which help in identification, verification and reporting of experimental results.
- SDS–PAGE therefore serve as a fundamental analytical technique used by biochemists, molecular biologists, and microbiologists for protein study, since it’s reliable, simple and economical.
Principle of SDS-PAGE
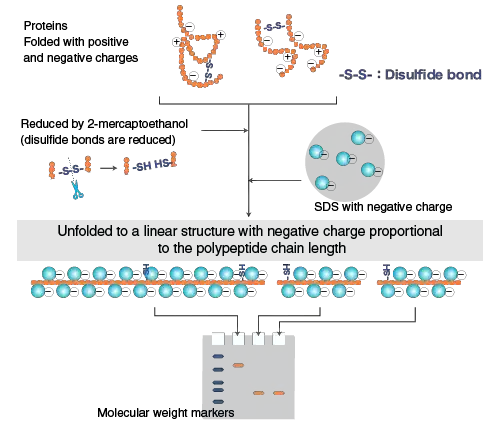
SDS–PAGE refers as Sodium Dodecyl Sulfate–PolyAcrylamide Gel Electrophoresis, and its main principle based on separation of proteins mainly by their molecular weight rather than charge or shape.
In this technique the detergent SDS (C₁₂H₂₅SO₄Na) bind strongly with proteins in ratio about 1.4 g SDS per 1 g protein, giving each molecule a uniform negative charge.
By this binding, native charge of protein is completely masked, so difference in electrophoretic mobility become dependent only by size of polypeptide chain.
SDS also cause denaturation of proteins, unfolding them into extended linear forms, thus eliminating influence of tertiary or quaternary structure during migration.
Proteins are then electrophoresed by a polyacrylamide gel, which act as a molecular sieve / matrix; small proteins migrate faster while large ones move slower due to pore resistance.
The movement occur when an electric field is applied, negatively charged protein–SDS complexes travel toward the positive electrode (anode).
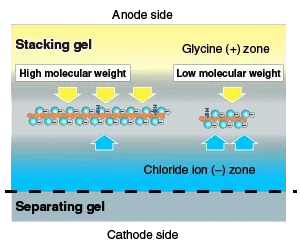
The gel usually consist of two layers – stacking gel (lower % acrylamide, low pH) and resolving gel (higher % acrylamide, higher pH).
In stacking gel, proteins are concentrated into thin sharp bands because of difference in ionic mobility between glycine, chloride, and protein ions, creating a moving boundary effect.
When sample reach the resolving gel, the uniform pH and higher acrylamide content cause proteins to separate strictly according to molecular size.
The rate of migration (Rf value) is inversely proportional to the logarithm of molecular weight, which allow estimation of unknown protein size using standard marker proteins.
The gel concentration can be varied depending on range of protein molecular weight studied, for example 7.5–12 % for typical protein separation.
During electrophoresis buffer (usually Tris–Glycine–SDS) maintain constant pH and ionic strength, ensuring stable current and accurate separation.
After run completion, proteins are stained by dyes like Coomassie Brilliant Blue or Silver stain, which bind to amino acids and make visible colored bands.
The principle therefore relies on concept that SDS equalize charge-to-mass ratio, so electrophoretic mobility is function mainly of molecular size within gel pores.
Hence, SDS–PAGE act as powerful analytical tool for determining molecular weight, purity and polypeptide composition of protein samples, and it’s still widely applied in biochemical analysis today.
What is the Role of SDS in SDS-PAGE?
SDS (Sodium Dodecyl Sulfate) act as an anionic detergent which play main role in denaturation and imparting uniform negative charge to proteins.
The SDS molecules bind non-covalently to the polypeptide backbone, approximately at ratio of 1.4 g SDS per 1 g protein, forming protein–SDS complex.
By this binding, native charges of amino acids are completely masked, so each protein acquires nearly same charge-to-mass ratio.
Because of that, during electrophoresis migration of proteins depend mainly by molecular weight, not by their original charge or shape.
SDS cause unfolding of proteins, disrupting non–covalent bonds like hydrogen, hydrophobic and ionic interactions which hold tertiary structure.
In presence of reducing agents like β–mercaptoethanol or DTT, disulfide linkages between subunits are broken, leading to full linearization of polypeptides.
The detergent convert globular proteins into flexible rod-like structures which move uniformly by gel pores under electric field.
SDS also help to maintain proteins in soluble form by preventing aggregation or precipitation during electrophoresis process.
By providing consistent charge per unit mass, SDS make comparison of migration distances between proteins meaningful and accurate for molecular weight estimation.
Without SDS, proteins would migrate according to their native charge and shape, causing overlapping and inaccurate separation.
Therefore SDS act as key component that standardize electrophoretic behavior of proteins in SDS–PAGE, ensuring reliable and reproducible resolution.
Its role can describe shortly as – denaturing agent, charge equalizer, and solubilizing compound all at once which makes the technique sturdy and widely used in protein analysis.
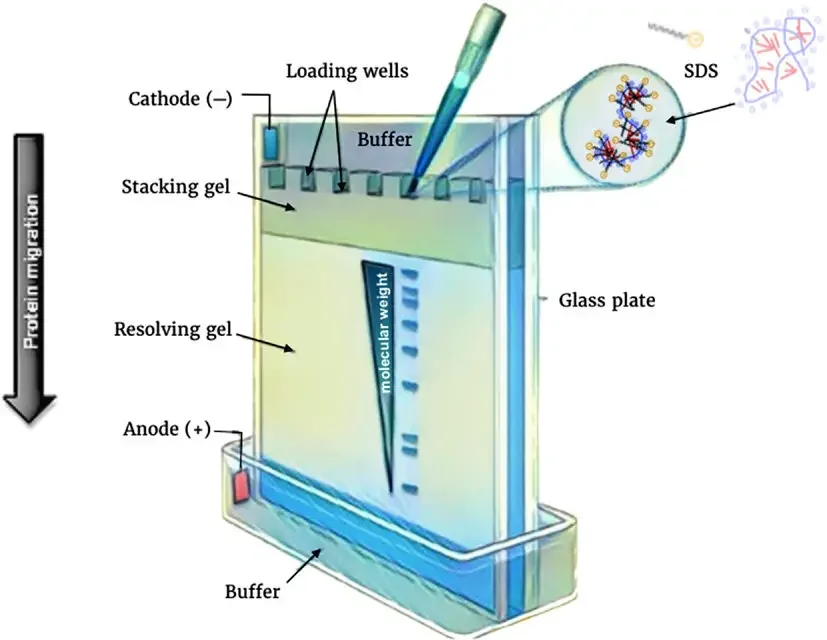
Requirement for SDS-PAGE and their roles
SDS (Sodium Dodecyl Sulfate)– It act as an anionic detergent which denature proteins by breaking non–covalent bonds, giving them uniform negative charge to ensure separation based mainly on size.
Polyacrylamide Gel – The gel serves as molecular sieve / matrix, where proteins are separated according to molecular weight. Smaller proteins migrate faster than larger ones through the pores.
Acrylamide and Bis–acrylamide – These two compounds form cross–linked network during polymerization. Acrylamide control pore size, while bis–acrylamide act as cross–linker for gel stability.
APS (Ammonium Persulfate)– It act as free radical initiator that start polymerization of acrylamide monomers, making the gel solidify.
TEMED (N,N,N′,N′-Tetramethylethylenediamine)– This compound accelerate formation of free radicals from APS, thus speed up polymerization process and allow uniform gel formation.
Tris–HCl Buffer– It maintain stable pH condition during electrophoresis (usually pH 6.8 for stacking gel and pH 8.8 for resolving gel) to preserve protein charge uniformity.
Glycine– Present in running buffer; it help in formation of ion front between chloride and glycine ions, which stack proteins into sharp bands before entering resolving gel.
Sample Buffer – It contain SDS, Tris–HCl, glycerol, β–mercaptoethanol / DTT, and bromophenol blue. It denature, reduce and give density to sample for loading convenience.
β–mercaptoethanol / DTT – They act as reducing agents that break disulfide bonds between cysteine residues, ensuring complete linearization of polypeptides.
Glycerol – It increase density of sample solution so it sink properly into gel wells during loading and not float away in buffer.
Bromophenol Blue – It used as tracking dye which migrate ahead of proteins to indicate electrophoresis progress visually.
Electrophoresis Buffer (Tris–Glycine–SDS)– Provide ions for conductivity and maintain constant electric field during run, while SDS in buffer sustain protein charge.
Power Supply Unit – Provide controlled electric current that move negatively charged protein–SDS complexes toward positive electrode.
Gel Casting and Electrophoresis Apparatus – It hold gels in vertical position, prevent leakage, and allow proper current flow through buffer chambers.
Staining Solution (Coomassie Brilliant Blue / Silver Stain) – Used for visualization of separated protein bands after electrophoresis.
Destaining Solution– Remove excess dye from gel background to make protein bands clearly visible.
Chemical Requirement for SDS PAGE
- Acrylamide/Bisacrylamide Solution 30% (29:1)
- 2.5X Tris-SDS Buffer (pH 8.8) 65 ml 125 ml 2-8o
- 5X Tris-SDS Buffer (pH 6.8)
- Prestained Protein Ladder
- 5X Tris-Glycine-SDS Gel Running Buffer
- 5X Sample Loading Buffer
- Protein Sample 1
- Protein Sample 2
- Staining solution
- Destaining solution
- Ammonium persulphate (APS)
- Tetramethylethylenediamine (TEMED)
- Agarose
Other Material Requireds;
- Glass wares: Conical flask, Measuring cylinder, Beaker
- Reagents: Distilled water
- Other requirements: Protein Electrophoresis apparatus, Micropipettes, Tips, Microwave/Burner/Hotplate
SDS or sodium dodecyl sulphate
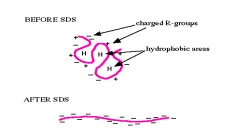
SDS or sodium dodecyl sulphate
SDS or Sodium Dodecyl Sulfate refers as an anionic detergent, widely used in biochemical and molecular biology techniques like SDS–PAGE for protein denaturation and solubilization.
It’s also called sodium lauryl sulfate (SLS) sometimes, and it’s a synthetic organic compound with chemical formula C₁₂H₂₅SO₄Na.
Structurally it composed of two distinct parts – a long hydrophobic dodecyl chain (C₁₂H₂₅–) and a hydrophilic sulfate group (–OSO₃⁻Na⁺).
Because of this dual nature, it act as amphipathic molecule, meaning it has both water-attracting (polar) and water-repelling (nonpolar) portions.
The hydrophobic tail interact with nonpolar regions of proteins, while the polar head interact with water, forming stable protein–SDS complexes.
SDS molecules align around hydrophobic protein surfaces causing disruption of internal hydrogen bonds and hydrophobic interactions which maintain native structure.
As result, proteins are unfolded into extended linear chains, completely denatured and surrounded by SDS molecules carrying uniform negative charge.
The binding ratio generally remain constant, around 1.4 g SDS per 1 g protein, which ensure equal charge distribution among different proteins.
In aqueous medium SDS form micelles above its critical micelle concentration (CMC ≈ 8 mM), where tails cluster inward and heads face outward, this property help in solubilizing hydrophobic compounds.
Its molecular weight is about 288.38 g/mol, and appear as white crystalline powder, soluble easily in water and ethanol but less in nonpolar solvents.
It’s strong surfactant, hence often used in cleaning agents, shampoos, and laboratory reagents though in biochemical context it act mainly as denaturant.
The sulfate group dissociate in solution, leaving negatively charged ions that interact electrostatically with protein backbone giving them uniform charge-to-mass ratio.
In SDS–PAGE, this property make separation depend mainly by molecular size, not charge or shape, which is key principle of the technique.
SDS is also known for causing foaming because of its surface activity; though in electrophoresis setup excessive foaming must be avoided for accuracy.
Hence, Sodium Dodecyl Sulfate act as crucial chemical reagent that bring denaturation, uniform charge and solubilization together – all essential for accurate protein electrophoretic analysis.

Protocol of SDS-PAGE
- Make sure that glass panels are secured to the unit, along with the spacers positioned on two edges.
- Make 1 % agarose (0.05g within 5ml distillated water). Boil until the agarose dissolves and then pour an even layer of horizontal agarose at the bottom of the plates to secure the plate. Allow it to solidify it to cool for 5-10 mins.
Preparation of 12% Separating Gel
For the preparation of separating gel include the ingredients as follows:
| Ingredients | Amount |
| 30% Acrylamide-bisacrylamide Solution | 6 ml |
| Distilled water | 3 ml |
| 2.5X Tris-SDS Buffer (pH 6.8) | 6 ml |
| 10% APS Solution | 125 ul |
| TEMED | 18 ul |
Place the gel between the plates and let it set for about one hour. After the gel has cooled, immediately pour it onto the plates.
Poured, add distilled water until it is level the gel. After about an hour, remove all the liquid by flipping the casting.
Preparation of 5% Stacking Gel
To prepare the stacking gel mix the ingredients in the following order:
| Ingredients | Amount |
| 30% Acrylamide-bisacrylamide Solution | 1.3 ml |
| Distilled water | 5.1 ml |
| 5X Tris-SDS Buffer (pH 6.8) | 1.6 ml |
| 10% APS Solution | 75 ul |
| TEMED | 10 ul |
After the addition of TEMED thoroughly mix all ingredients by swirling the beaker. Place the stacking gel on the top of the separating gel. Then, immediately, place the comb, making sure to avoid air bubbles. It should be allowed to set for about 30 minutes.
Important: Acrylamide is a potential neurotoxin, and must be handled with extreme attention. Always wear a face-mask and wear gloves.
- Place 1X Tris-Glycine-SDS Gel Running Buffer into the unit in a way that it connects the two electrodes and consequently completes that flow. Take the comb out of the stacking Gel cautiously.
- Sample Preparation: Take 2 tubes for protein samples. Label them accordingly. You should take 20 ml of each sample from the tube and add 5ml of the 5X sample loading buffer it. The tubes that contain Protein Samples at 100oC in the bath of boiling water. Don’t boil the tube that has PrestainedProtein Ladder.
- Take 5ml of the Prestained Protein Ladder and 20ml of the samples right after the heat treatment of the wells made by the comb within the Stacking Gel.
- The power cable should be connected to an electrophoretic power source according to the standard protocols: Red-Anode as well as Black-Cathode. The electrophoresis process is 100 V at 10 mA until the dye front is 0.5 cm higher than that of the gel sealing.
- Take the gel out between the plates by putting it into the plastic tray that contains distillate water. The gel should be cleaned for 1 minute. Then, rinse the gel with water and go to destain the stain procedure.
Staining and Destaining of Gel
- After removing the gel from the electrophoresis unit, remove any excess water from the gel’s surface.
- Place the gel in a tray or container and add 50ml of Staining Solution to cover the gel completely. Allow the gel to soak in the staining solution until the protein bands become apparent. Sometimes, it may be necessary to incubate the gel in the staining solution overnight to ensure optimal band visualization.
- Once the desired staining intensity is achieved, carefully remove the gel from the staining solution. The staining solution can be reused for up to three times, as long as it remains effective.
- Rinse the stained gel with distilled water to remove the excess stain. Gently shake the gel in a tray filled with distilled water, and then replace the water with fresh distilled water. Repeat this rinsing process three to four times, changing the water each time, to ensure thorough removal of the stain.
- Prepare 50ml of Destaining Solution and add it to the tray containing the gel. The destaining solution is typically a solvent that helps remove the background stain from the gel. Shake the tray containing the gel at a constant rate to facilitate the destaining process.
- Continue destaining until the protein bands on the gel become clear and distinct, while the background stain is removed. This may take some time and may require periodic evaluation of the gel’s appearance.
- Once the desired destaining is achieved, carefully remove the gel from the destaining solution. Similar to the staining solution, the destaining solution can also be reused for up to three times, provided it still effectively removes the background stain.
After completing the staining and destaining steps, the gel can be visually examined to observe the protein bands. Alternatively, the gel can be further processed for documentation, such as by scanning or photographing, to preserve and analyze the results.
Observation and Result of SDS-PAGE
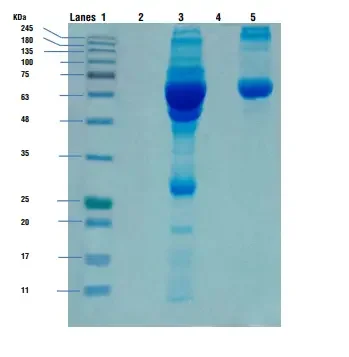
- Lane 1: Prestained Protein Ladder
- Lane 3: Protein Sample 1
- Lane 5: Protein Sample 2
Interpretation
Following staining, and then destaining, gel, you can compare the molecular mass of the samples to the weight of the protein marker. Protein Sample 1 is a serum sample so multiple bands are observed. Protein Sample 2 is a purified protein and therefore 1 major band is visible.
Sample Preparation for SDS-PAGE
- Take 1ml of culture solution of E.coli strain DH5α, in 1.5 ml of eppendorf’s tube.
- Then centrifuge at 12000 rpm for 5 min.
- Then discard the supernatant.
- Then add 500 μl of TE in the tube and dissolve the tube by gentle shaking.
- Then add same volume of sample buffer.
- Finally heat the sample on boiling bath for 5 minutes and then immediately keep on
ice.
The sample to analyze is optionally mixed with a chemical denaturant if so desired, usually SDS for
proteins or urea for nucleic acids. SDS is an anionic detergent that denatures secondary and non–
disulphide linked tertiary structures, and additionally applies a negative charge to each protein in
proportion to its mass. Urea breaks the hydrogen bonds between the base pairs of the nucleic acid,
causing the constituent strands to separate. Heating the samples to at least 60 °C further promotes denaturation.
Protocol of SDS-PAGE
The SDS-PAGE method is composed of gel preparation, sample preparation, electrophoresis, protein staining or western blotting and analysis of the generated banding pattern.
1. Gel Preparation
Gel preparation is a crucial step in gel electrophoresis for separating proteins based on their size. Here are the steps involved in gel preparation:
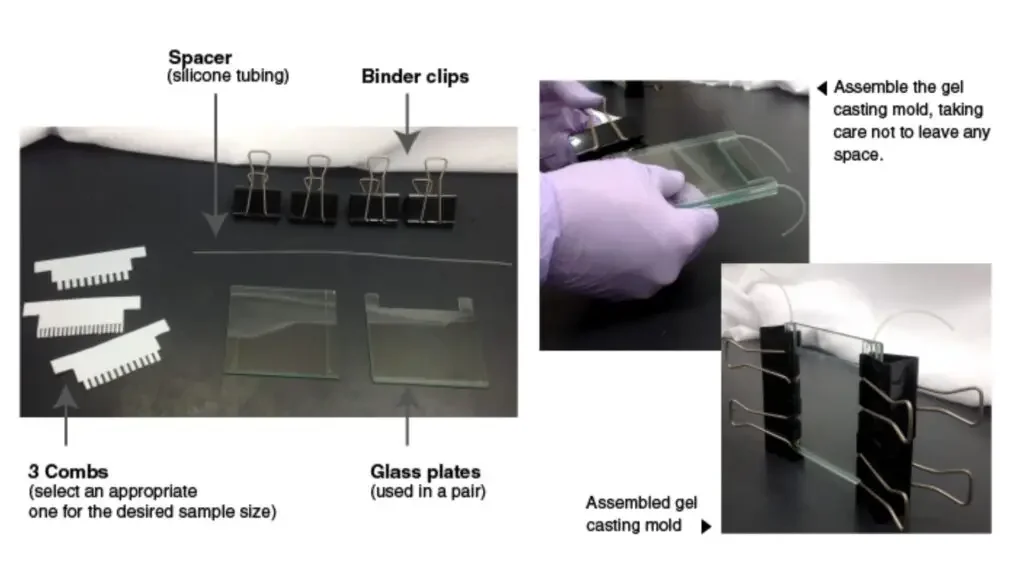
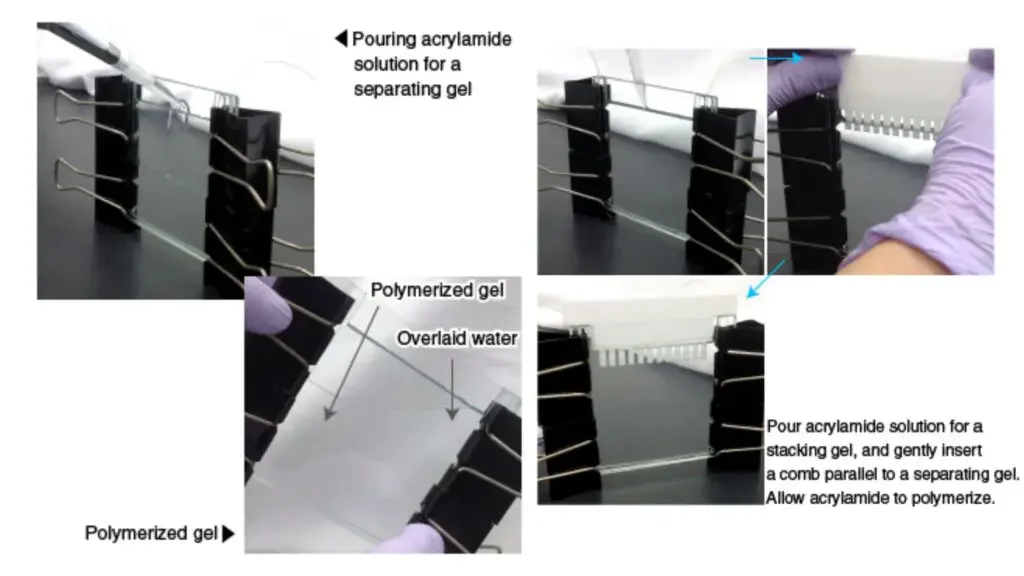
- Gathering Materials: Gather all the necessary materials, including combs, glass plates, spacer (silicone tubing), and binder clips. Ensure that the glass plates are thoroughly cleaned with ethanol before use.
- Assembling the Gel Casting Mold: Assemble the gel casting mold using the cleaned glass plates. The mold consists of two glass plates held together with binder clips, leaving a space between them to pour the gel.
- Preparing the Acrylamide Solution: Combine all the reagents required for gel preparation, except for TEMED (Tetramethylethylenediamine). This solution usually includes acrylamide, bisacrylamide, buffer, and water.
- Adding TEMED: When the gel is ready to be poured, add TEMED to the acrylamide solution. TEMED acts as a catalyst for polymerization.
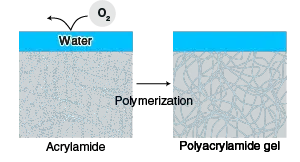
- Pouring the Separating Gel: Pour the acrylamide solution into the space between the glass plates, forming the separating gel. To prevent contact with air (oxygen), which inhibits polymerization, overlay the solution with water. Allow the acrylamide to polymerize for 20-30 minutes to form a gel.
- Removing Air Bubbles: Before the gel polymerizes, add butanol to the top of the gel. The addition of butanol helps to remove any unwanted air bubbles that may be present in the gel.
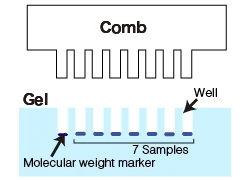
- Inserting the Comb: Insert a comb into the spaces between the glass plates. The comb is used to create wells or lanes in the gel, where the protein samples will be loaded. Choose an appropriate comb with the desired number of lanes based on the number of samples and molecular weight markers to be used.
- Gel Polymerization: Allow the acrylamide to polymerize completely, forming a solid gel. Once polymerized, the gel becomes a “gel cassette” held between the glass plates.
Gel preparation is tailored to the specific requirements of the experiment. The concentration of acrylamide determines the mesh size of the gel, affecting the separation of proteins. Higher acrylamide concentrations (6-15%) are suitable for separating small proteins, while gradient gels with varying acrylamide concentrations may be used for specialized applications.
In addition, a stacking gel can be used before the separating gel to concentrate proteins before they enter the separation process. The stacking gel takes advantage of the differential migration rates of glycine ions, chloride ions, and proteins to create a concentrated protein zone.
Proper gel preparation ensures the optimal conditions for separating proteins based on their size during gel electrophoresis.
Once the gel is prepared, it can be used for gel electrophoresis experiments to separate and analyze proteins based on their molecular weight or other properties.
2. Sample preparation
Sample preparation is a crucial step in protein analysis and gel electrophoresis. Here are the steps involved in sample preparation:
- Boiling Water: Start by boiling water in a beaker. This water will be used for various steps in the sample preparation process.
- Preparation of Sample Buffer: Add 2-mercaptoethanol to the sample buffer. 2-mercaptoethanol helps to denature the proteins and reduce any disulfide bonds that may be present.
- Sample and Marker Preparation: Place the buffer solution in microcentrifuge tubes and add the protein sample to it. Similarly, prepare separate tubes containing molecular weight markers. These markers help determine the molecular weights of the proteins during gel electrophoresis.
- Protein Denaturation: Boil the samples, including the protein sample and markers, for a short duration, typically less than 5 minutes. This step ensures complete denaturation of the proteins.
- Addition of Sample Buffer: After boiling, add an appropriate volume of sample buffer to each sample tube. The sample buffer helps to stabilize the proteins and provides them with the necessary charge for electrophoresis.
- Mixing and Heat Treatment: Mix the contents of the tubes by flicking them gently to ensure proper mixing of the sample and buffer. Heat the tubes at 100°C for 3 minutes using a heat block. This step further denatures the proteins and helps to break down any secondary or tertiary structures.
- Centrifugation: After heat treatment, centrifuge the tubes at high speed (around 15,000 rpm) for 1 minute at 4°C. This step helps to separate any precipitated or insoluble material, and the supernatant is used for SDS-PAGE (sodium dodecyl sulfate-polyacrylamide gel electrophoresis).
- Loading Buffer Addition: To the protein sample (cell or tissue lysate), add an equal volume of loading buffer. The loading buffer contains additional components such as tracking dyes and stabilizers, which aid in sample loading and visualization during electrophoresis.
- Additional Boiling and Centrifugation: Boil the mixture of protein sample and loading buffer at 95°C for 5 minutes. Following this, centrifuge the mixture at a high speed (around 16,000 xg) for 5 minutes. These steps ensure complete denaturation and proper mixing of the sample before gel electrophoresis.
- Storage or Electrophoresis: The prepared samples can be stored at -20°C if not immediately used for gel electrophoresis. Alternatively, proceed with gel electrophoresis by loading the samples onto the gel.
Proper sample preparation is essential for accurate and reliable results in gel electrophoresis. It involves denaturation, reduction of disulfide bonds, and appropriate mixing of samples with buffer and loading dye to ensure optimal separation and analysis of proteins.
3. Electrophoresis
The steps of electrophoresis involve loading the denatured samples onto a polyacrylamide gel, applying a voltage to induce migration of proteins, and observing the separated bands. Here are the detailed steps:
- Gel Preparation: Prepare the polyacrylamide gel by casting it in a gel cassette or gel apparatus. The gel should be immersed in an electrophoresis buffer containing suitable electrolytes.
- Sample Loading: Using a pipette, carefully load the denatured protein samples into the wells of the gel. Each sample is placed in its designated well.
- Power Supply Connection: Place the gel cassette with the loaded samples into the electrode assembly of the electrophoresis apparatus. Ensure that it is securely fixed in the clamp stand. Connect the electrophoresis unit to a power supply.
- Electrophoresis Buffer Addition: Pour 1x electrophoresis buffer into the opening of the casting frame, allowing it to fill the wells of the gel. The buffer acts as a medium for the migration of proteins during electrophoresis.
- Application of Voltage: Once the gel is ready and the buffer is added, cover the tank with a lid to create a closed system. Turn on the power supply and apply a suitable voltage, typically around 100 V or 10-20 V per cm of gel length. The voltage causes negatively charged molecules, including proteins, to migrate through the gel towards the positively charged anode.
- Electrophoresis Duration: Allow the electrophoresis process to run for the desired duration, typically ranging from half an hour to several hours depending on factors such as the voltage applied and the length of the gel. The separation occurs as proteins of different sizes migrate through the gel at different rates, based on their molecular weights.
- Visualization of Bands: After electrophoresis, the gel is removed from the apparatus and bands are visualized. One common method is to use UV light to observe the protein bands. UV illumination allows for the visualization of bands that may be stained or labeled with fluorescent dyes.
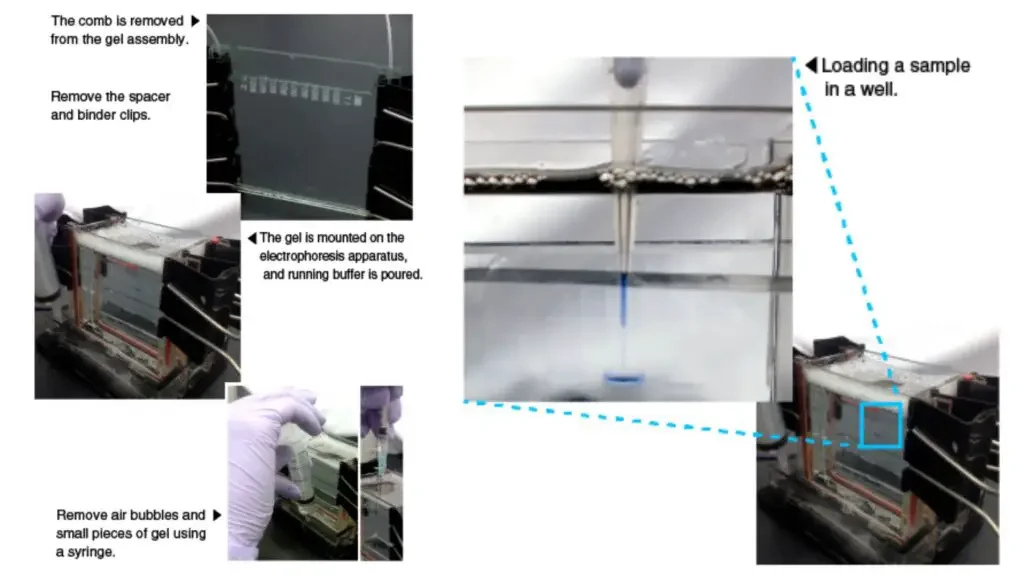
These steps provide a general overview of the process of electrophoresis, allowing proteins to be separated based on their molecular sizes. It is important to note that specific protocols and variations may exist depending on the type of electrophoresis technique or analysis being performed.
4. Gel staining
Gel staining is a crucial step in protein analysis that allows for the visualization and detection of separated proteins within the gel. Here is an overview of the gel staining procedure:
- Fixation: After the completion of electrophoretic separation, the gel is typically fixed to immobilize the proteins and prepare them for staining. A common fixation solution consists of 50% ethanol and 10% glacial acetic acid. The gel is immersed in this solution for approximately 1 hour.
- Staining solution preparation: While the gel is being fixed, the staining solution is prepared. For Coomassie staining, a widely used and straightforward method, the staining solution is composed of 50% methanol, 10% glacial acetic acid, and 0.1% Coomassie Brilliant Blue dye. The staining solution should be mixed thoroughly.
- Staining: After the fixation period, the gel is transferred to a fresh staining solution, ensuring complete immersion of the gel. The staining step allows the dye to bind to the proteins, resulting in their visualization as blue bands. The gel is typically stained for a period ranging from 1 to 12 hours, although the specific staining time can vary depending on the desired intensity of staining and the sensitivity of the proteins being analyzed.
- Destaining: Once the desired level of staining has been achieved, the gel is destained to remove excess dye and enhance the contrast between the stained protein bands and the gel background. The destaining solution consists of 40% methanol and 10% glacial acetic acid. The gel is immersed in the destaining solution, and the solution is changed several times to facilitate efficient destaining. The destaining process continues until the background is clear, and the protein bands are distinctly visible.
Different staining methods and dyes may have variations in the staining and destaining procedures. Some staining techniques, such as silver staining, amido black staining, and fast green FCF staining, may require different staining solutions and destaining protocols. Additionally, fluorescent stains and immunological detection methods like Western Blotting offer alternative approaches for protein visualization and analysis.
Gel staining is a critical step in protein analysis as it allows researchers to observe and analyze the separated proteins within the gel, providing valuable insights into protein size, abundance, and potential modifications.
5. Analysis
Analysis of protein samples following gel electrophoresis involves the examination and interpretation of the banding pattern obtained through protein staining. Here are some important considerations during the analysis:
- Banding Pattern- Protein staining in the gel results in the formation of distinct bands representing different proteins based on their molecular weight. The banding pattern provides a visual representation of the proteins present in the sample and their relative abundance.
- Glycoproteins: Glycoproteins, which have varying levels of glycosylation, can exhibit broader and blurred bands on the gel. This is due to the uneven adsorption of SDS at the glycosylation sites, affecting the migration and resulting in less well-defined bands.
- Membrane Proteins: Membrane proteins, characterized by their hydrophobic nature and tendency to bind lipids, may have lower solubility in aqueous solutions. This can lead to precipitation and incomplete migration in SDS-PAGE, causing a “tailing” effect above the band of the transmembrane protein. Adjusting the SDS concentration and protein loading can help mitigate this issue.
- Overloading: Overloading the gel with a high concentration of a particular soluble protein can result in a semicircular band, which may obscure or overlap with neighboring bands. This can make it challenging to distinguish proteins with similar molecular weights.
- Contrast and Purity: The contrast between bands within a lane can provide insights into the purity of the protein sample. Low contrast may indicate the presence of multiple proteins (low purity) or, in the case of purified proteins, proteolytic degradation. Degradation can be observed as degradation bands below the intact protein band, eventually resulting in a homogeneous “smear” below the degraded band.
- Documentation: The banding pattern is typically documented by photographing or scanning the gel. This allows for a permanent record of the protein separation and facilitates further analysis and comparison. Documentation is important for subsequent recovery or identification of specific protein bands.
- Gel Extraction: In some cases, it may be necessary to recover the proteins present in individual bands for further analysis or downstream applications. Gel extraction techniques can be employed to isolate and collect proteins from specific gel regions, enabling subsequent characterization or identification.
The analysis of protein banding patterns obtained from gel electrophoresis provides valuable information about the protein samples, including their molecular weight, relative abundance, and potential modifications. By carefully examining and interpreting these patterns, researchers can gain insights into the composition and behavior of the proteins under investigation.
Molecular mass determination
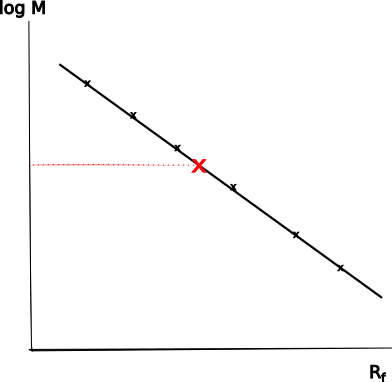
Molecular mass determination is an important aspect of protein analysis following gel electrophoresis. The following steps are involved in the process:
- Measurement of Relative Migration Distances- To determine the molecular weight of proteins accurately, the relative migration distances of individual protein bands are measured within the separating gel. These measurements are typically conducted in triplicate to improve precision.
- Calculation of Relative Mobility (Rf value)-The relative mobility, also known as the Rf value or Rm value, is calculated by dividing the distance traveled by the protein band by the distance traveled by the buffer front. Both distances are measured from the beginning of the separation gel. The buffer front distance corresponds approximately to the migration distance of a dye, such as bromophenol blue, present in the sample buffer.
- Construction of a Semi-logarithmic Plot – The relative distances of proteins from the size marker are plotted semi-logarithmically against their known molecular weights. This plot allows for a linear relationship to be observed between the logarithm of the molecular weight and the relative mobility.
- Determination of Molecular Weight- By comparing the relative mobility of an unknown protein with the linear portion of the plot or performing regression analysis, the molecular weight of the protein can be estimated. This estimation is based on its relative mobility value.
Factors Affecting Molecular Weight Determination:
- Glycosylated Proteins- Protein bands with glycosylations may appear blurred, making the determination of their molecular weight less precise.
- Basic or Acidic Proteins- Proteins containing a high number of basic amino acids, such as histones, may exhibit slower migration due to their positive charges. This can result in an overestimation of their molecular weight or even prevent their migration into the gel. Conversely, proteins rich in acidic amino acids may migrate more quickly, leading to an underestimation of their molecular mass.
Molecular mass determination is a valuable tool for characterizing proteins and assessing their sizes. By accurately measuring relative migration distances and utilizing size markers, researchers can estimate the molecular weight of unknown proteins, providing insights into their structural properties and facilitating further analysis and comparison.
Different Gel Staining Methods
1. Classical & colloidal Coomassie staining –
The method is widely used because it is simple, low‐cost and effective for lots of proteins.
The dye (eg Coomassie Brilliant Blue R-250 or G-250) binds non‐specifically to proteins (via basic amino acids and aromatic residues) so that bands become visible as blue against a clear background.
The sensitivity is moderate (detection limit ~100 ng per band for classical Coomassie) so low‐abundance proteins may not be seen.
A variant labelled as “colloidal Coomassie” uses G-250 in special buffer and gives higher sensitivity (~10 ng) and better reproducibility, but cost is higher.
One drawback: the background may remain if destaining is incomplete, so gel fixation and proper destaining must be done.
2. Silver staining –
A more sensitive method than Coomassie for detecting proteins of low abundance.
The principle: silver ions react (reduce) in presence of proteins (often with sulfhydryl or carboxyl groups) to form visible metallic silver deposits on the protein bands.
Detection limits can reach ~2-5 ng per band (or even lower) depending on protocol.
However proteins stained by silver often cannot be easily used for downstream applications (eg mass spectrometry) because silver can interfere.
Also the technique is more time-consuming and more prone to variability/artefacts (eg negative staining of some proteins).
3. Fluorescent / specialized staining methods–
For higher sensitivity or compatibility with downstream analytic like MS or imaging.
For example: dyes like SYPRO Orange or other fluorescent stains may allow detection of lower amounts (nanogram level) and broader dynamic range.
Imaging may use near-infrared fluorescence after Coomassie or fluorescent stains to further boost sensitivity.
Drawbacks: usually cost is higher, equipment (fluorescent scanner) may be needed, and some stains may require special handling or are less compatible with other downstream steps.
4. Other less common / alternative stains–
Methods like zinc based negative staining, or rapid microwave‐assisted protocols, or co‐electrophoretic stains have also been developed. arXiv
When choosing a stain the trade-off is often sensitivity vs cost/time vs downstream compatibility (eg mass spec, transfer to blot).
Summary of main comparison:
- Coomassie – easiest, moderate sensitivity, good for general use.
- Silver – high sensitivity, more complex, limited downstream uses.
- Fluorescent/special – very high sensitivity and dynamic range, higher cost/complexity.
Electrophoresis Buffers
Electrophoresis Buffers
| Electrophoretic Method | |||
|---|---|---|---|
| SDS-PAGE (InvitrogenTM) | BN-PAGE (InvitrogenTM) | NSDS-PAGE | |
| Sample Buffer | 106 mM Tris HCl 141 mM Tris Base 0.51 mM EDTA 0.22 mM SERVA Blue G-250 0.175 mM Phenol Red 2% LDS 10% Glycerol pH 8.5 | 50 mM BisTris 50 mM NaCl 16mMHCl 10% Glycerol 0.001% Ponceau S pH 7.2 | 100 mM Tris HCl 150 mM Tris Base 0.01875% Coomassie G-250 0.00625% Phenol Red 10% Glycerol pH 8.5 |
| Run Buffer | 50 mM MOPS 50 mM Tris Base 1 mM EDTA 0.1% SDS pH 7.7 | Cathode 50 mM BisTris 50 mM Tricine 0.02% Coomassie G-250 pH 6.8 Anode 50 mM BisTris 50 mM Tricine pH 6.8 | 50 mM MOPS 50 mM Tris Base 0.0375% SDS pH 7.7 |
Applications of SDS-PAGE
- Determining molecular weight of proteins is a common use. The mobility of unknown proteins is compared with standard markers to estimate size.
- Assessing protein purity / homogeneity is done by observing whether a single sharp band or multiple bands are present, indicating contaminants or degradation.
- Analysis of subunit composition of multi‐subunit proteins is possible when they are reduced and run on SDS-PAGE, showing individual polypeptide chains.
- Monitoring protein expression levels (for example in recombinant systems) is carried out by loading samples from different conditions, and comparing band intensities.
- Investigation of post‐translational modifications (PTMs) such as phosphorylation or glycosylation is possible because the modified protein may shift mobility (migrate differently) on the gel.
- Use in downstream techniques: SDS-PAGE is used prior to Western blotting (to transfer proteins from gel) and for sample preparation for mass spectrometry.
- Comparative analysis of polypeptide composition between different samples (for example different tissues, organisms, treatments) is enabled by SDS-PAGE, since the pattern of bands shows differences in composition.
- Clinical/diagnostic applications: For example the separation of viral proteins (like in an HIV test) or evaluation of proteinuria (urine proteins) use SDS-PAGE.
- Food / agricultural research: The technique is applied for analysing protein composition in foods (milk, cheese, yogurt) and this goes in food science & tech.
Advantages of SDS PAGE
- High resolution in protein separation is offered by SDS-PAGE, because the proteins (denatured by SDS) are separated mainly by size, which makes band patterns clearer and more distinct.
- Good reproducibility is achieved since the method is well‐standardised and many labs will get similar results if protocol is followed, so comparative studies are easier and more reliable.
- Versatility is provided: many types of proteins (soluble, membrane, nuclear) can be run by SDS-PAGE, and it is compatible with downstream techniques (like blotting).
- Uniform charge/mass ratio is created by SDS binding to proteins, which means that migration is less influenced by protein native charge/shape and mainly by molecular weight, so interpretation is simpler and more consistent.
- Cost‐effectiveness is another advantage because basic equipment and reagents are required and many labs already have the apparatus, so the barrier to using SDS-PAGE is relatively low.
- Ease of use and adaptability: The procedure has been widely used for many years, it is well documented, and it integrates with other analytic steps (eg gel staining, western blot, MS) so the method is handy and flexible.
- Good chemical/physical stability of the gel matrix (polyacrylamide) means that the separation system is sturdy and hardy, which aids in consistent performance and fewer failures from gel breakdown.
Limitations of SDS PAGE
- The native conformation of proteins is lost when SDS-PAGE is used, so functional studies (like enzyme activity or protein-protein binding) cannot be done reliably.
- Only separation by size / molecular weight is effectively offered; separation by other properties (charge, isoelectric point, hydrophobicity) is very limited.
- Extremely large proteins or very small proteins may not be resolved well: large ones may migrate too slowly or get stuck, small ones may run off or overlap.
- Quantitative accuracy is limited: band intensity gives only rough estimate of protein amount and many factors (loading, staining, destaining) affect it.
- Sample preparation and gel casting are time-consuming, require care (to avoid leaks, ensure correct polymerisation), and the gels cannot be reused which makes throughput lower.
- Aggregates or protein complexes may behave unpredictably (migrate irregularly or smear) so interpretation becomes tricky, especially for samples that are complex or multi‐component.
- The technique sometimes uses toxic monomers (eg acrylamide) and handling risks / safety concerns need to be considered.
- Because it is primarily qualitative or semi-quantitative, for precise quantification or high throughput you may need more advanced / automated methods.
FAQ
What is SDS-PAGE?
SDS-PAGE (Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis) is a technique used to separate proteins based on their molecular weight. It involves denaturing proteins with SDS and subjecting them to electrophoresis through a polyacrylamide gel matrix.
Why is SDS used in SDS-PAGE?
SDS is an anionic detergent that denatures proteins, coats them with a negative charge relative to their mass, and unfolds them into linear structures. This allows the proteins to be separated based on size during electrophoresis.
What is the purpose of a stacking gel in SDS-PAGE?
The stacking gel serves as a sieving matrix that helps concentrate the protein samples into thin, focused bands before they enter the separating gel. It improves the resolution and separation of proteins during electrophoresis.
How does SDS-PAGE separate proteins?
During SDS-PAGE, an electric field is applied to the gel, causing proteins to migrate through the gel matrix based on their size. The smaller proteins move faster and migrate further, while larger proteins migrate more slowly and remain closer to the sample loading wells.
What is the purpose of a molecular weight marker in SDS-PAGE?
A molecular weight marker, also known as a protein ladder, is a mixture of proteins with known molecular weights. It is loaded onto the gel alongside the samples and serves as a reference to estimate the size of the unknown proteins in the samples.
How is protein visualization achieved in SDS-PAGE?
After electrophoresis, proteins are often visualized by staining the gel with dyes such as Coomassie Brilliant Blue or silver stain. These dyes bind to proteins and allow their detection as bands or spots on the gel.
Can SDS-PAGE be used to determine protein concentration?
While SDS-PAGE provides information about protein size and separation, it is not a reliable method for quantifying protein concentration. Spectrophotometric methods, such as Bradford assay or BCA assay, are commonly used for protein quantification.
Can SDS-PAGE identify post-translational modifications?
SDS-PAGE alone cannot identify specific post-translational modifications (PTMs). Additional techniques, such as Western blotting or mass spectrometry, are typically employed to detect and characterize PTMs.
What are the limitations of SDS-PAGE?
Some limitations of SDS-PAGE include its inability to provide detailed structural information, difficulty in visualizing nucleic acids, limited assessment of purity, variability in results, and restricted dynamic range for very large or small proteins.
What are the applications of SDS-PAGE?
SDS-PAGE is widely used in various fields of research and industry. It is utilized for protein analysis, purification, characterization, and protein-protein interaction studies. It is also an important technique in molecular biology, biochemistry, biotechnology, and clinical diagnostics.
- Nowakowski AB, Wobig WJ, Petering DH. Native SDS-PAGE: high resolution electrophoretic separation of proteins with retention of native properties including bound metal ions. Metallomics. 2014 May;6(5):1068-78. doi: 10.1039/c4mt00033a. PMID: 24686569; PMCID: PMC4517606.
- https://ruo.mbl.co.jp/bio/e/support/method/sds-page.html
- https://www.aatbio.com/resources/buffer-preparations-and-recipes/sds-page-10x-sds-running-buffer
- https://www.bio-rad.com/en-in/applications-technologies/sds-page-analysis?ID=LW7FGX4VY
- http://microbiology.ucdavis.edu/heyer/wordpress/wp-content/uploads/2013/11/dna_page.pdf
- https://www.cbrnetechindex.com/Biological-Detection/Technology-BD/Electrophoresis-BD-T/Polyacrylamide-Gel-Electrophoresis-BD-E
- https://www.rockland.com/resources/sds-page-protocol/
- https://www.bio.davidson.edu/Courses/genomics/method/SDSPAGE/SDSPAGE.html
- https://www.ruf.rice.edu/~bioslabs/studies/sds-page/gellab2.html
- https://www.sigmaaldrich.com/IN/en/technical-documents/protocol/protein-biology/gel-electrophoresis/sds-page
- https://thesciencenotes.com/sds-page/