What is Active Site of an Enzyme?
- The active site of an enzyme is a crucial region where substrate molecules bind and undergo a chemical reaction. It plays a central role in catalyzing the reaction by facilitating the binding of the substrate and reducing the activation energy required for the reaction to occur. The active site is often described as a small cleft or cavity within the enzyme, composed of approximately 10-15 amino acid residues.
- The primary function of the active site is twofold: binding activity and catalytic activity. The binding activity refers to the ability of the active site to enhance the affinity between the enzyme and the substrate, ensuring a strong and specific interaction. This specificity is essential for the enzyme’s function, as it allows it to selectively bind and act upon a particular substrate. The amino acid residues surrounding the active site help to hold the substrate molecule in the correct position during biochemical reactions, further enhancing the binding affinity.
- The catalytic activity of the active site is responsible for the actual chemical transformation of the substrate into the product. Within the active site, there are specific residues that catalyze the reaction by participating in the formation and breaking of chemical bonds. These catalytic residues can stabilize the transition state of the substrate, leading to the formation of the desired product. By reducing the activation energy required for the reaction, the active site enables a higher rate of reaction and efficient conversion of substrates.
- The active site is strategically located deep within the enzyme’s structure, resembling a hole or a small depression. Its position within the enzyme ensures that the substrate molecule is brought into close proximity with the catalytic residues, allowing for efficient enzymatic activity. Although the active site occupies a relatively small portion of the enzyme’s volume, usually around 10-20%, it is the most critical region as it directly catalyzes the chemical reaction.
- The active site’s specificity and catalytic efficiency are a result of its unique molecular arrangement and structure. Each active site is finely tuned and optimized to bind a specific substrate and catalyze a particular reaction. The interactions between the active site and the substrate are highly specific and rely on complementary shapes, charge distributions, and other molecular properties.
- In some cases, enzymes may require additional cofactors to perform their function effectively. These cofactors can bind to the active site, assisting in the catalytic process by providing chemical groups or facilitating electron transfers.
- Overall, the active site of an enzyme is a specialized region that enables the enzyme to bind and act upon specific substrates, catalyzing the conversion of substrates into products. Its binding and catalytic activities are essential for the enzyme’s function, allowing for highly efficient and specific biochemical reactions.
Definition of Enzyme’s Active Site
The active site of an enzyme is a small region within the enzyme where substrate molecules bind and undergo a chemical reaction, leading to the formation of products.
What is Binding site?
- The binding site refers to a specific region within the active site of an enzyme where the substrate molecule binds. It plays a crucial role in orienting the substrate for catalysis and facilitating the formation of an enzyme-substrate complex.
- The interaction between the binding site and the substrate is non-covalent and transient, initially. Several types of interactions contribute to the binding of the substrate, including hydrogen bonds, van der Waals interactions, hydrophobic interactions, and electrostatic forces. These interactions ensure that the substrate is held in a defined orientation within the active site.
- The charge distribution on both the substrate and the binding site must be complementary, allowing for a balanced distribution of positive and negative charges. This prevents repulsive forces and promotes a stable interaction between the substrate and the enzyme. The binding site typically consists of non-polar amino acids, although polar amino acids can also be present.
- For successful binding and selectivity, at least three contact points between the substrate and the binding site are required. These contacts contribute to the stereo-, regio-, and enantioselectivity of the enzyme-substrate interaction. Each contact point corresponds to a specific part of the substrate molecule that interacts with the binding site.
- Proper protein folding and maintenance of the enzyme’s tertiary structure are essential for the binding site to function correctly. Various interactions between amino acid residues stabilize the enzyme’s structure. Disruptions to these interactions, such as extreme pH, high temperature, or high ion concentrations, can cause the enzyme to denature and lose its catalytic activity.
- A tighter fit between the binding site and the substrate generally leads to increased reaction efficiency. It is believed that enhancing the tightness between the binding site of an enzyme, such as DNA polymerase, and its substrate can improve the fidelity or accuracy of the enzymatic reaction.
- Most enzymes have deeply buried active sites that can be accessed by the substrate through specific access channels. These channels allow the substrate molecule to reach the binding site within the enzyme.
- There are three proposed models for enzyme-substrate binding: the lock and key model, the induced fit model, and the conformational selection model. These models describe different ways in which enzymes and substrates interact. The induced fit and conformational selection models are not mutually exclusive, and a protein may exhibit characteristics of both models. Factors like temperature can influence the binding pathway, with higher temperatures favoring conformational selection over induced fit.
- In summary, the binding site is a region within the active site of an enzyme that specifically interacts with the substrate, facilitating the formation of an enzyme-substrate complex and initiating the catalytic reaction.
Features that Determine Active Site Specificity
The specificity of the active site in enzymes, which allows them to bind to specific substrates, is determined by various features. These features influence the binding affinity and selectivity of the active site for different substrates. Some of the key features that determine active site specificity include:
- Size and shape: The active site of an enzyme is structured in a way that it can accommodate and interact specifically with certain substrates. The size and shape of the active site determine which substrates can fit into it properly. The complementary shape ensures a strong interaction between the enzyme and its specific substrate.
- Polarity or non-polarity: Enzyme active sites may contain polar or non-polar amino acids. Polar molecules tend to interact with other polar molecules, while non-polar molecules prefer interactions with other non-polar molecules. The presence of specific polar or non-polar amino acids in the active site can attract or repel different parts of the substrate, creating a better fit and enhancing specificity.
- Charge distribution: The active site may have amino acids with positive or negative charges. Opposite charges attract each other, facilitating substrate binding, while similar charges repel each other. The charge distribution within the active site enables the attraction of substrates or specific regions of substrates while repelling others, contributing to substrate specificity.
- Hydrophobicity or hydrophilicity: Enzyme active sites can possess hydrophobic or hydrophilic amino acids. Hydrophobic amino acids tend to interact with other hydrophobic molecules, while hydrophilic amino acids prefer interactions with hydrophilic substrates. This property aids in the selective binding of substrates that have compatible hydrophobic or hydrophilic characteristics.
- Co-factor requirements: Some enzymes require co-factors, such as vitamins or minerals, to bind effectively to their substrates. Co-factors play a crucial role in enhancing the specificity of the active site by assisting in substrate recognition and binding. For example, certain B vitamins serve as co-factors for enzymes involved in energy production, thereby influencing the substrate specificity of those enzymes.
By combining these various features, the active site of an enzyme is finely tuned to bind specific substrates while excluding others. The unique sequence, structure, and physical properties of the active site allow enzymes to exhibit high specificity, ensuring that they catalyze specific reactions with precision.
Reaction Mechanism of an Enzyme With Example
The reaction mechanism of an enzyme involves the binding of a specific substrate to the active site, the formation of an enzyme-substrate complex, catalytic conversion of the substrate into products, and the release of the products. Let’s delve deeper into this mechanism using the example of sucrase and sucrose:
- Substrate binding: Sucrose, the substrate, attaches to the active site of the enzyme sucrase. The active site of sucrase is specifically shaped to accommodate the sucrose molecule, forming an enzyme-substrate complex (E-S complex).
- Catalytic reaction: Once the sucrose is bound to the active site, a catalytic reaction occurs. This reaction involves the conversion of sucrose into its constituent molecules, glucose and fructose. The sucrase enzyme facilitates this conversion by altering the structural conformation of the sucrose molecule, leading to the formation of a transition state.
- Transition state formation: The transition state represents an intermediate state during the reaction where the bonds within the substrate are in the process of being broken and rearranged. This structural change enables the conversion of the substrate into products.
- Product formation: The transition state of sucrose eventually leads to the formation of the enzyme-product complex (E-P complex). In this complex, the sucrase enzyme is still bound to the products, glucose and fructose.
- Product release: Finally, the products, glucose and fructose, are released from the active site of the sucrase enzyme, leaving it free to catalyze further reactions.
This example illustrates how sucrase, an enzyme with a specific active site for sucrose, catalyzes the breakdown of sucrose into glucose and fructose. The binding of the substrate, the conversion of the substrate into products through a transition state, and the subsequent release of the products are key steps in the reaction mechanism of the enzyme.
It is important to note that enzymes are not consumed or altered during the reaction. They can repeatedly bind to substrates, catalyze reactions, and release products, making them highly efficient catalysts in biological processes.
Important Characteristics of Active site
The characteristics of an active site that are important for its function include:
- Hydrophobicity: The initial binding between the substrate and enzyme occurs through non-covalent bonds, including hydrophobic interactions. Hydrophobic binding enhances the binding affinity of the substrate to the active site. Other types of interactions, such as van der Waals forces, hydrogen bonds, and electrostatic forces, also contribute to the formation of the enzyme-substrate complex.
- Flexibility: Active sites exhibit flexibility, allowing them to undergo conformational changes to facilitate the conversion of substrates into products. This flexibility enables the active site to accommodate different substrates and adapt to the reaction conditions.
- Reactivity: The active site of an enzyme is reactive and catalyzes specific reactions. The reactivity of the active site depends on various environmental factors, such as temperature, pH, and concentrations of the enzyme and substrate. The active site binds to the substrate, lowering the activation energy required for the reaction and promoting catalysis.
- Net charge: The active site primarily consists of non-polar amino acid residues, which carry no charge or have a net charge of zero. However, some active sites may also contain polar amino acids with positive or negative charges. The net charge of the active site determines the amino acids that bind to the enzyme. Complementary pairing between the active site and the substrate is essential for the formation of the enzyme-substrate complex. Repulsion may occur if there is the same charge on both the catalytic site and the substrate.
These characteristics of the active site contribute to its ability to bind specific substrates, catalyze reactions, and provide a favorable environment for the conversion of substrates into products. The unique properties of the active site play a crucial role in the high specificity and efficiency of enzymatic reactions.
Role of Active Site
The active site of an enzyme plays a crucial role in facilitating and accelerating chemical reactions. It performs two major functions: substrate binding and catalytic activity. Let’s explore the role of the active site in these two conditions.
First condition:
In the absence of an enzyme catalyst, the substrate requires a higher activation energy to undergo the transition from its initial state to the transition state. This transition state is represented as “St” and involves a change in the substrate’s conformation. Eventually, the substrate reaches the transition state and releases a product (P). This process requires a significant amount of energy.
Second condition:
In the presence of an enzyme catalyst, the substrate binds to the active site of the enzyme. The enzyme acts as a catalyst, facilitating the reaction between the substrate and the transition state. The enzyme modifies the substrate and guides it towards the transition state, represented as “ESt.” At the transition state, the enzyme and substrate undergo a reaction, leading to a change in the substrate’s configuration. This change results in the formation of an enzyme-product complex, which eventually releases the product (P).
The active site of an enzyme significantly reduces the activation energy required for the reaction to proceed. By lowering the activation energy, the enzyme increases the rate of the reaction. The energy level of the substrate is higher than that of the product but lower than the transition state of the substrate. The active site of the enzyme provides a favorable environment and facilitates the interaction between the substrate and the transition state, promoting the conversion of substrate into product.
In summary, the active site of an enzyme plays a critical role in catalyzing reactions by lowering the activation energy and increasing the rate of the reaction. It provides a specific binding site for the substrate and creates an optimal environment for the reaction to occur efficiently. The active site acts as a catalyst, mediating the conversion of substrate into product and enabling the enzyme to perform its biological function.
Active Site Binding Theories
There are two hypotheses regarding how an enzyme’s active site binds to substrates. The lock and key model and the induced fit model are examples.
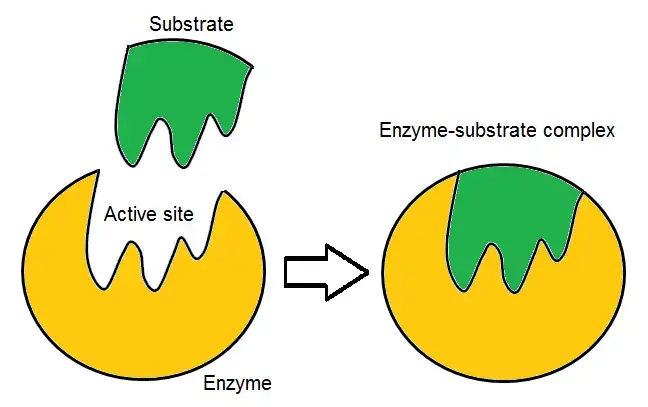
1. The Lock and Key Model
- The Lock and Key Model is a concept proposed by Emil Fischer, a chemist from the 19th century, to explain the specificity of enzyme-substrate interactions. According to this model, the active site of an enzyme is a specific and rigid structure that perfectly complements the shape and chemical properties of its substrate, just like a lock and key fit together.
- In this model, the active site and substrate are considered as pre-existing structures that do not require any further modification to interact with each other. When the substrate comes into contact with the active site, it fits precisely into the active site’s shape, forming an enzyme-substrate complex. The interactions between the active site and the substrate are strong and specific, allowing for high catalytic efficiency.
- However, as more research has been conducted, limitations of the Lock and Key Model have become apparent. For example, certain competitive enzyme inhibitors can bind tightly to the active site but do not undergo the expected catalytic reactions. This observation challenges the Lock and Key Model’s prediction of high catalytic efficiency based solely on tight binding. Similarly, non-competitive inhibitors that do not bind to the active site but still affect the enzyme’s activity cannot be explained by this model.
- Despite its limitations, the Lock and Key Model provides a valuable conceptual framework for understanding enzyme-substrate interactions. It highlights the importance of the complementary shape and chemical properties between the active site and the substrate for efficient catalysis. Over time, further refinements to this model, such as the Induced Fit Model and the Conformational Selection Model, have been proposed to better explain the complexity of enzyme-substrate binding and catalysis.
2. The Induced Fit Model
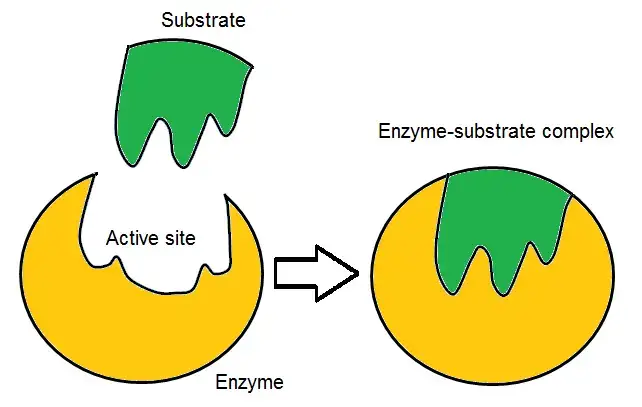
- The Induced Fit Model, proposed by Daniel Koshland, expands upon the Lock and Key Model to provide a more comprehensive understanding of enzyme-substrate interactions. According to this model, the active site of an enzyme and the substrate are initially not an exact fit for each other in their resting states.
- In the Induced Fit Model, the active site of the enzyme is flexible and can undergo conformational changes to accommodate the substrate. It is like a glove that changes its shape to fit the hand. The enzyme’s initial conformation has some affinity for the substrate, but it is not a perfect match. As the substrate approaches the enzyme, interactions between the binding site and the substrate induce changes in their shapes.
- The induced conformational changes in the active site and the substrate enable a tighter and more specific binding. This conformational adjustment is crucial for the catalytic activity of the enzyme. It allows for optimal positioning of the substrate within the active site, facilitating the catalytic reaction. The active site and the substrate continue to interact dynamically during the catalytic process.
- Once the reaction is complete and the product is formed, the product and the enzyme are no longer compatible. They separate from each other, and the active site returns to its initial shape to accommodate new substrate molecules.
- The Induced Fit Model is supported by experimental observations, such as the movement of protein domains during catalysis and the creation of microenvironments within the active site that promote catalytic activity. It provides a more dynamic and flexible explanation for enzyme-substrate interactions compared to the rigid Lock and Key Model.
- Overall, the Induced Fit Model highlights the dynamic nature of enzyme-substrate binding, where both the enzyme and the substrate undergo changes to achieve optimal interaction and catalysis. This model is widely accepted in the scientific community due to its compatibility with experimental evidence.
3. Conformational selection hypothesis
- The Conformational Selection Hypothesis is a model that describes the mechanism of substrate binding to enzymes based on the concept of conformational flexibility. According to this hypothesis, enzymes exist in multiple conformational states, and only certain conformations are capable of effectively binding to a substrate.
- Enzymes are dynamic molecules that can adopt different shapes or conformations. These conformations may vary in their structural arrangement and accessibility of the active site. In the absence of a substrate, enzymes exist in equilibrium between different conformational states.
- The Conformational Selection Hypothesis suggests that when a substrate molecule is present, it selectively binds to the enzyme’s active site, inducing a conformational change in the enzyme. This binding event stabilizes the enzyme-substrate complex and shifts the equilibrium of the conformational ensemble towards conformations that are favorable for substrate binding.
- In other words, the enzyme already possesses the range of conformations necessary for substrate binding, and the substrate selectively binds to the specific conformation that is complementary to its shape and chemical properties. The binding process is driven by the inherent flexibility of the enzyme and the specific molecular recognition between the enzyme and substrate.
- This hypothesis suggests that enzymes sample different conformations in solution, and only when a substrate molecule encounters the enzyme in the appropriate conformational state, binding occurs. The bound substrate then undergoes catalytic reactions within the active site, leading to the formation of products.
- The Conformational Selection Hypothesis emphasizes the importance of conformational dynamics and the role of pre-existing enzyme conformations in substrate binding and catalysis. It provides a framework to understand how enzymes can selectively recognize and bind to specific substrates from a pool of potential molecules.
- Overall, the Conformational Selection Hypothesis highlights the dynamic nature of enzymes and their ability to adopt different conformations to accommodate substrate binding. This model helps explain how enzymes achieve specificity and efficiency in their catalytic functions.
Types of non-covalent interactions in Active Site
Non-covalent interactions play a crucial role in various biological processes, including enzyme-substrate interactions within the active site. Here are the different types of non-covalent interactions:
- Electrostatic interactions: Oppositely charged groups, such as positively charged amino acid side chains and negatively charged substrates, attract each other through electrostatic interactions. These interactions are mediated by the attraction between ions of opposite charge, contributing to the stability of enzyme-substrate complexes.
- Hydrogen bonds: Hydrogen bonds occur when a partially positive hydrogen atom interacts with a partially negative electron donor group, typically containing oxygen, nitrogen, or fluorine. Hydrogen bonds are directional and can provide both structural stability and specificity in enzyme-substrate interactions.
- Van der Waals forces: Van der Waals forces arise from transient fluctuations in electron distribution, leading to attractive forces between oppositely charged groups. Van der Waals forces include two components: dispersion forces (arising from temporary fluctuations in electron distribution) and dipole-dipole interactions (arising from permanent dipoles). Although individually weak, these forces collectively contribute to the overall stability of enzyme-substrate complexes.
- Hydrophobic interactions: Hydrophobic interactions involve the tendency of non-polar hydrophobic groups to aggregate together and minimize contact with polar solvents, such as water. In the context of enzyme active sites, hydrophobic interactions can contribute to substrate binding by facilitating the burial of hydrophobic groups within the enzyme’s interior, away from the surrounding aqueous environment.
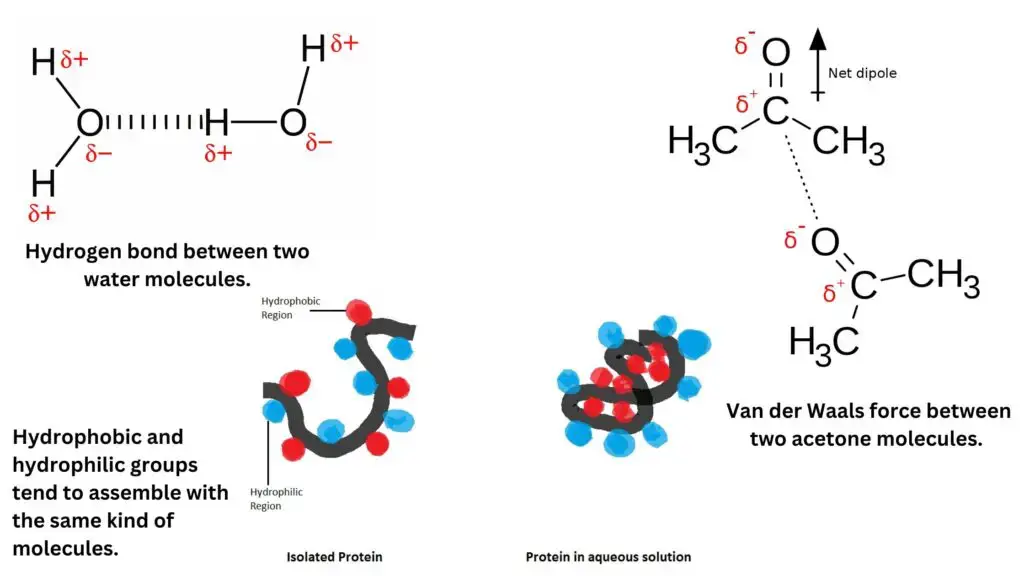
These non-covalent interactions collectively contribute to the formation and stabilization of enzyme-substrate complexes. By providing specificity, stability, and flexibility, these interactions enable enzymes to selectively bind substrates and facilitate catalysis. The strength and significance of each interaction type depend on factors such as the nature of the interacting groups, their spatial arrangement, and the surrounding environment. Understanding these non-covalent interactions is essential for comprehending enzyme-substrate recognition and the overall functioning of biological systems.
Catalytic process Mechanisms
Catalytic process mechanisms are the various ways in which enzymes facilitate and accelerate chemical reactions. Several mechanisms have been identified that explain the role of enzymes in catalysis. Here are the key mechanisms involved:
- Approximation of the reactant: Enzymes bring the substrate and active site into close proximity, increasing the effective concentration of the substrate and reducing the energy required for them to interact. This process also minimizes the involvement of solvent molecules and allows the active site to reorient the substrate, facilitating the reaction.
- Covalent catalysis: Some enzymes form transient covalent bonds with substrates, lowering the activation energy required for the reaction. This involves the formation and subsequent breakdown of covalent bonds between the enzyme and substrate. Examples include enzymes like proteases and kinases.
- Nucleophilic catalysis: Enzymes with nucleophilic groups donate electrons to substrates, forming covalent bonds during the transition state. The strength of this interaction depends on the nucleophilic group’s ability to donate electrons and the electrophile’s ability to accept them. Amino acids such as serine, cysteine, aspartate, and glutamine can act as nucleophiles.
- Electrophilic catalysis: In this mechanism, amino acids in the active site of the enzyme act as electrophiles, while the substrates act as nucleophiles. Similar to nucleophilic catalysis, this process involves the formation of covalent bonds during the transition state.
- Metal ions: Metal ions can play various roles in catalysis. They can bind to negatively charged substrate groups, enhance electrophilicity, bridge between the active site and substrate, and induce conformational changes that favor the reaction.
- Acid/base catalysis: Protons and hydroxide ions can act as specific acid or base catalysts in some reactions. In general acid/base catalysis, groups in the substrate and active site function as Brønsted-Lowry acids and bases. They stabilize the nucleophile or electrophile by providing positive or negative charges. Amino acids like aspartate, glutamate, histidine, and cysteine can act as general acids or bases.
- Conformational distortion: This mechanism suggests that both the active site and the substrate can undergo conformational changes to achieve optimal fit and facilitate the reaction. It helps explain the accelerated reaction rates observed in enzymatic reactions that cannot be fully explained by other mechanisms.
- Preorganized active site complementarity to the transition state: This theory suggests that the active site of an enzyme is preprogrammed to bind specifically to the transition state of the substrate rather than the ground state. By minimizing the energy required to rearrange solvent molecules, the active site enhances the electrostatic complementarity with the transition state, facilitating the reaction.
These different catalytic mechanisms highlight the diverse strategies employed by enzymes to enhance the rates of chemical reactions and perform their biological functions. Enzymes often combine multiple mechanisms to achieve efficient catalysis.
| Catalytic Process Mechanism | Description |
|---|---|
| Approximation of the reactant | Enzymes bring the substrate and active site into close proximity, increasing effective concentration and reducing the desolvation energy required for the reaction. The active site may reorient the substrate to reduce activation energy. Binding is favored by entropy, as solvent molecules are excluded from the active site. The active site can manipulate the substrate’s molecular orbital for suitable orientation. |
| Covalent catalysis | Enzymes form transient covalent bonds with substrates, lowering activation energy. This involves two steps: formation and breakdown of covalent bonds. The former is the rate-limiting step, while the latter regenerates the intact enzyme. Examples include proteases, kinases, and phosphatases. |
| Nucleophilic catalysis | Enzyme’s nucleophile donates electrons to the substrate, forming a covalent bond during the transition state. The strength of interaction depends on the nucleophilic group’s ability to donate electrons and the electrophile’s ability to accept them. Amino acids such as serine, cysteine, aspartate, and glutamine can act as nucleophiles. |
| Electrophilic catalysis | Amino acids in the active site act as electrophiles, while the substrate acts as a nucleophile. The mechanism is similar to nucleophilic catalysis but with reversed roles. Cofactors are often required as amino acid side chains may not be strong enough in attracting electrons. |
| Metal ions | Metal ions have multiple roles in catalysis, such as binding to negatively charged substrate groups, increasing electrophilicity, bridging between the active site and substrate, and inducing conformational changes. |
| Acid/base catalysis | Protons and hydroxide ions may directly act as acids and bases in specific acid/base catalysis. General acid/base theory involves groups in the substrate and active site acting as Brønsted-Lowry acids and bases. They stabilize the nucleophile or electrophile formed during catalysis by providing positive or negative charges. Amino acids like aspartate, glutamate, histidine, and cysteine can serve as acids or bases. |
| Conformational distortion | Enzymatic reactions sometimes show acceleration beyond existing theories. Conformational distortion suggests that both the active site and substrate can undergo conformational changes to fit each other. |
| Preorganized active site complementarity to the transition state | The active site is preprogrammed to bind perfectly to the substrate in the transition state, minimizing the energy required to rearrange solvent molecules. Charged groups within the active site attract substrates, ensuring electrostatic complementarity. |
Catalysis mechanisms Examples
Catalysis Mechanisms of Glutathione reductase
Glutathione reductase (GR) plays a crucial role in the catalytic process of regenerating glutathione (GSH) by breaking the disulfide bond in the oxidized form of glutathione (GSSG). Here is an overview of the catalytic mechanisms involved in GR:
- GR Structure and Cofactors: Glutathione reductase is a dimeric enzyme composed of two identical subunits. It requires two cofactors, namely one NADP (nicotinamide adenine dinucleotide phosphate) and one FAD (flavin adenine dinucleotide). The active site, responsible for catalysis, is located at the interface between the two subunits. Within the active site, there are two cysteine residues and the FAD cofactor.
- Reduction of FAD: The catalytic process begins with the reduction of FAD by NADPH, resulting in the formation of FADH-. This reduction process involves the transfer of one electron from NADPH to FAD, converting it into its reduced form.
- Attack on Disulfide Bond: FADH-, acting as a nucleophile, attacks the disulfide bond formed between two cysteine residues. This attack leads to the formation of a single SH bond and a sulfur anion (S^-) group. The sulfur anion group will act as a nucleophile in the subsequent steps.
- Breaking the Disulfide Bond in GSSG: The sulfur anion (S^-) group generated from the previous step attacks the disulfide bond in the oxidized form of glutathione (GSSG). This attack results in the cleavage of the disulfide bond, forming a cysteine-SG complex, where SG represents the glutathione molecule.
- Release of the First SG^- Anion: The first SG^- anion is released from the cysteine-SG complex. It receives a proton from an adjacent SH group and from the first glutathione monomer, leading to the formation of a cysteine-SH complex.
- Attack on the Cysteine-SG Complex: The adjacent sulfur anion (S^-) group, generated from another cysteine residue, now attacks the disulfide bond within the cysteine-SG complex. This attack results in the release of the second SG^- anion.
- Formation of Glutathione Monomers: The released second SG^- anion receives a proton from the solution, ultimately forming the second glutathione monomer.
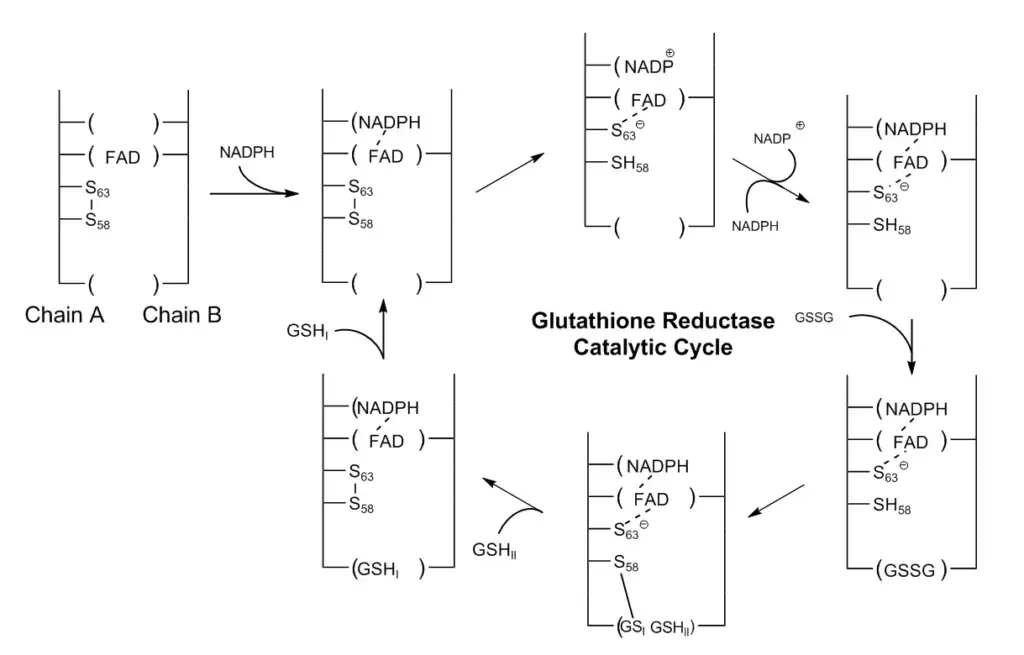
By following these catalytic steps, glutathione reductase facilitates the regeneration of reduced glutathione (GSH) by breaking the disulfide bond in the oxidized form (GSSG). This process allows glutathione to continue its vital role in protecting cells from oxidative damage caused by reactive oxygen species.
Catalysis Mechanisms of Chymotrypsin
Chymotrypsin, a serine endopeptidase found in pancreatic juice, plays a crucial role in protein and peptide hydrolysis. The catalytic mechanism of chymotrypsin involves a catalytic triad consisting of three amino acid residues in its active site: Ser-195, His-57, and Asp-102. Here is an overview of the catalysis mechanisms of chymotrypsin:
- Nucleophilic Attack: The first phase of chymotrypsin catalysis involves Ser-195 acting as a nucleophile and attacking the peptide bond carbon in the substrate. This nucleophilic attack forms a tetrahedral intermediate. The nucleophilicity of Ser-195 is enhanced by His-57, which abstracts a proton from Ser-195. This deprotonation is facilitated by the negatively charged carboxylate group (RCOO-) in Asp-102. Additionally, the tetrahedral oxyanion intermediate formed in this step is stabilized by hydrogen bonds from Ser-195 and Gly-193.
- Protonation and Leaving Group Formation: In the second stage, His-57 acts as a base to protonate the leaving group (R’NH) of the substrate, resulting in the formation of R’NH2. The leaving group then dissociates from the intermediate, leaving behind the acylated Ser-195. Subsequently, His-57 acts as a base again, abstracting a proton from a water molecule.
- Nucleophilic Attack and Cleavage: The hydroxide anion (OH-) generated by the deprotonation of water acts as a nucleophile and attacks the acyl-enzyme complex. This attack leads to the formation of a second tetrahedral oxyanion intermediate. Similar to the previous step, the tetrahedral oxyanion intermediate is stabilized by hydrogen bonds.
- Cleavage and Regeneration: Ser-195 leaves the tetrahedral intermediate, resulting in the cleavage of the carbon-oxygen (C-O) bond between the enzyme and the peptide substrate. A proton is transferred from His-57 to Ser-195, restoring all three amino acids (Ser-195, His-57, and Asp-102) to their initial states.
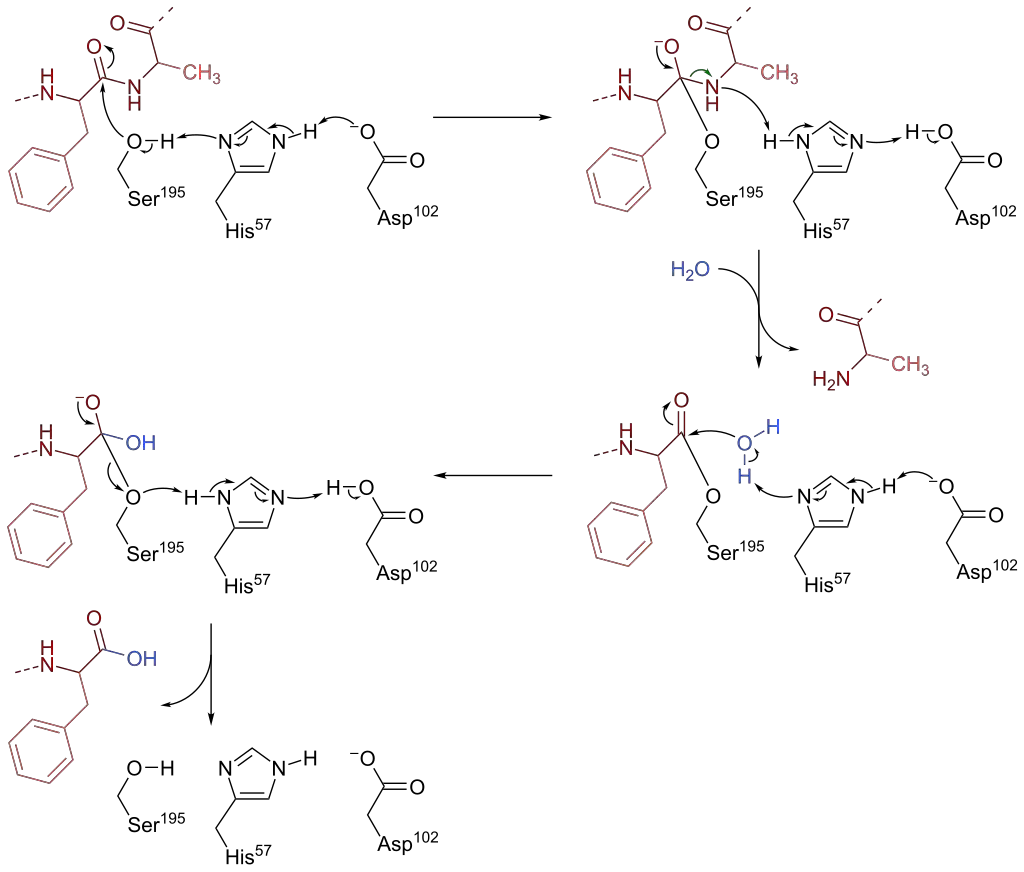
Through these catalytic steps, chymotrypsin facilitates the hydrolysis of peptide bonds, specifically targeting L-isomers of tyrosine, phenylalanine, and tryptophan. The catalytic triad plays a crucial role in enhancing the nucleophilicity of Ser-195 and stabilizing the reaction intermediates, allowing chymotrypsin to efficiently carry out its proteolytic function.
Substrate Unbinding
The process of substrate unbinding refers to the dissociation of a substrate from the active site of an enzyme. Several factors influence the kinetics and dynamics of substrate unbinding. Here are some key factors that affect the unbinding process:
- Ligand Size: The size of the substrate can influence its residence time in the active site. Generally, larger ligands tend to stay bound to the enzyme for a longer duration compared to smaller ligands. This is attributed to the increased number of interactions and contacts between the larger substrate and the enzyme’s active site.
- Ligand Flexibility: The flexibility of a substrate also plays a role in its unbinding kinetics. Substrates with more rotatable bonds tend to have longer residence times in the active site. However, it’s worth noting that the correlation between ligand flexibility and residence time may be influenced by other factors, such as ligand size.
- Protein Flexibility: The flexibility of the protein itself can impact substrate unbinding. Less flexible proteins typically result in longer residence times for the substrate in the active site. This can be attributed to the restricted conformational changes required for the substrate to dissociate from the enzyme.
- Hydrogen Bonding: The presence of hydrogen bonds between the substrate and the enzyme can affect the unbinding process. Hydrogen bonds that are shielded from the surrounding solvent environment contribute to a stronger interaction between the substrate and the enzyme, reducing the likelihood of unbinding.
These factors collectively contribute to the overall kinetics of substrate unbinding. Understanding the factors that influence unbinding can provide insights into the enzymatic activity, specificity, and regulation, as well as the design of enzyme inhibitors or drug molecules that target specific enzymatic processes.
What are Cofactors?
Cofactors play a crucial role in the functioning of enzymes by assisting them in carrying out their catalytic activities. Here are some key points about cofactors:
- Definition and Types: Cofactors are non-protein molecules that bind to enzymes and aid in their catalytic function. They can be classified into various types, including coenzymes, metal ions, and prosthetic groups. Coenzymes, such as vitamins and ATP, are organic molecules that often participate in redox reactions and carry chemical groups or electrons between enzymes. Metal ions, such as iron or zinc, can act as electron carriers or participate in catalytic reactions. Prosthetic groups are tightly bound cofactors that are permanently attached to the enzyme.
- Binding and Interaction: Coenzymes typically bind to enzymes through non-covalent interactions, such as hydrogen bonds or hydrophobic interactions. However, in some cases, covalent bonds can form between the coenzyme and the enzyme. For example, the heme group in cytochrome C is bound to the protein through a thioester bond. Coenzymes can transiently associate with the enzyme during the reaction or remain permanently bound.
- Cofactor Function: Cofactors are essential for the proper functioning of enzymes. They can participate in various enzymatic reactions by accepting or donating electrons, transferring chemical groups, or providing structural support. For example, flavin coenzymes, such as flavin adenine dinucleotide (FAD) and flavin mononucleotide (FMN), have a distinct isoalloxazine ring system that allows them to accept one or two electrons. This property enables them to act as electron carriers and participate in oxidation-reduction reactions.
- Cofactor Role in Enzyme Activity: Enzymes that require a cofactor to function properly are called apoenzymes. Without their respective cofactors, apoenzymes are often inactive or exhibit reduced catalytic activity. The binding of the cofactor to the active site of the enzyme forms the holoenzyme, which is the active and functional form of the enzyme. The cofactor provides the necessary chemical groups or electrons required for the enzyme to carry out its specific catalytic reaction.
Cofactors are essential for the proper functioning of many enzymes, enabling them to carry out complex biochemical reactions. Their diverse chemical properties and roles make them crucial components in enzyme catalysis and regulation.
What are Inhibitors?
Inhibitors are substances that interfere with the normal interaction between enzymes and substrates, leading to a decrease in the rate of a chemical reaction. There are different types of inhibitors, including reversible and irreversible forms. Here are some key points about inhibitors:
- Competitive Inhibitors: Competitive inhibitors specifically target the free enzyme molecules and compete with substrates for binding to the active site. They often have structural similarities to the substrates or the enzyme-substrate (ES) complex, allowing them to fit into the active site and block substrate entry. Alternatively, they can induce conformational changes in the active site that prevent proper substrate binding. Competitive inhibitors can be overcome by increasing the substrate concentration, as higher substrate levels outcompete the inhibitors for binding to the active site.
- Non-competitive Inhibitors: Non-competitive inhibitors bind to both the free enzyme and the ES complex. Unlike competitive inhibitors, they do not compete for the active site. Instead, they bind to a different site on the enzyme, leading to a change in the enzyme’s three-dimensional structure. This structural alteration can block the entry or exit of substrates, thereby reducing the enzyme’s catalytic activity. Non-competitive inhibitors cannot be overcome by increasing substrate concentration since they do not directly interact with the active site.
- Irreversible Inhibitors: Irreversible inhibitors also bind to the active site of an enzyme but form covalent bonds with amino acid residues, resulting in a permanent alteration of the active site. These inhibitors typically contain electrophilic groups that react with nucleophilic amino acids in the active site, forming stable covalent adducts. Once bound, irreversible inhibitors cannot be easily displaced and permanently deactivate the enzyme. Examples of irreversible inhibitors include compounds like cyanide, which can bind to the active site and inhibit key enzymes involved in cellular respiration.
In summary, inhibitors can disrupt enzyme-substrate interactions and affect the rate of enzymatic reactions. Competitive inhibitors compete with substrates for the active site, non-competitive inhibitors bind to a different site and alter the enzyme’s structure, while irreversible inhibitors form covalent bonds with the active site, rendering the enzyme permanently inactive. Understanding the mechanisms of inhibition is essential in both basic research and drug development, as inhibitors can be targeted to modulate enzyme activity in various biological processes.
Examples of Enzymes
Enzymes play a vital role in catalyzing various chemical reactions in living organisms. Here are a few examples of enzymes and the reactions they catalyze:
- Maltase and Starch: Maltase is an enzyme that breaks down starch into simpler sugars. Starch, a complex carbohydrate, is hydrolyzed by maltase into individual sugar molecules. This reaction occurs when maltase cleaves the glycosidic bonds between sugar units in starch, resulting in the formation of smaller, sweeter-tasting sugars. Starch + Water → Sugars
- Pepsin and Protein: Pepsin is an enzyme involved in the digestion of proteins in the stomach. It works in a highly acidic environment and breaks down proteins into smaller peptide fragments. Pepsin catalyzes the hydrolysis of peptide bonds, which are the bonds that link amino acids in proteins. The reaction involves the cleavage of the peptide bonds by pepsin in the presence of stomach acid, leading to the production of amino acids. Protein + Acid → Amino acids
- DNA Polymerase: DNA polymerase is an enzyme responsible for DNA replication, a crucial process in cellular reproduction. It synthesizes new DNA strands by adding nucleotides in a complementary manner to the existing DNA template strand. DNA polymerase catalyzes the formation of phosphodiester bonds between adjacent nucleotides, resulting in the elongation of the DNA molecule. This synthesis reaction allows for the accurate replication of the genetic material during cell division. DNA triphosphate molecule + DNA strand → Longer DNA strand + diphosphate molecule
These examples highlight the diverse roles of enzymes in various biological processes. Enzymes facilitate the conversion of complex substances into simpler ones, enabling the efficient utilization of nutrients and the maintenance of essential cellular functions. Understanding the specific reactions catalyzed by enzymes provides insights into the fundamental processes underlying life.
Key Points about Active site
- An active site is the specific location within an enzyme where a substrate binds to initiate and catalyze a reaction. It is often referred to as the enzyme’s catalytic surface.
- The active site is formed by the arrangement of approximately 10-15 amino acid residues in the enzyme’s structure.
- The active site has a unique geometric shape and chemical properties that enable it to recognize and bind specifically to its corresponding substrate.
- The specificity of the active site allows only substrates with complementary shapes to bind, similar to a key fitting into a lock.
- The active site of an enzyme can catalyze various chemical and biological pathways, depending on the nature of the substrate.
- When the enzyme-substrate complex is formed, both the substrate and the active site undergo structural changes. This may involve bending or rearranging the bonds in the substrate molecule to facilitate its conversion into a product.
- Enzymes exhibit catalytic activity primarily because of their active sites. The active site’s properties, such as shape, size, charge, and stereospecificity, contribute to the catalysis of specific reactions.
- The active site of an enzyme plays a role in inducing the transition state of the substrate. The transition state refers to the intermediate state during a reaction where the bonds are being broken and formed, leading to the conversion of the substrate into products.
FAQ
What is an active site?
The active site of an enzyme is a specific region or pocket where the substrate molecule(s) bind and undergo a chemical reaction.
Why is the active site important?
The active site is crucial because it determines the enzyme’s specificity for its substrate(s) and facilitates the catalytic reaction by providing a favorable environment for the reaction to occur.
How is the active site formed?
The active site is formed by specific amino acid residues in the enzyme’s protein structure that create a unique three-dimensional shape and chemical environment.
Can multiple substrates bind to the active site simultaneously?
Yes, some enzymes have active sites capable of binding multiple substrate molecules simultaneously, facilitating reaction pathways involving multiple substrates.
How does the active site recognize its substrate?
The active site recognizes the substrate through various interactions, including hydrogen bonding, electrostatic interactions, and hydrophobic interactions, which complement the specific shape and chemical properties of the substrate.
Can the active site change its shape?
In some cases, the active site can undergo conformational changes to accommodate the substrate(s) more effectively, a phenomenon known as induced fit.
How does the active site lower the activation energy of a reaction?
The active site provides a suitable environment for the reaction, stabilizes transition states, and may even directly participate in the reaction by providing functional groups or metal ions to facilitate catalysis.
Can the active site be altered by enzyme mutations?
Yes, mutations in the gene encoding an enzyme can lead to changes in the amino acid sequence, potentially affecting the structure and function of the active site.
Can the active site be blocked by inhibitors?
Yes, inhibitors can bind to the active site, preventing the substrate from binding and inhibiting enzyme activity. This is the basis for some therapeutic drugs and chemical compounds used in research.
Can the active site be targeted for drug design?
Yes, understanding the structure and function of the active site can guide the design of drugs that specifically bind to the active site, either inhibiting or enhancing enzyme activity for therapeutic purposes.