Pulsed Field Gel Electrophoresis (PFGE) is a highly effective genotyping technique used for the separation and analysis of large DNA molecules, such as entire genomic DNA. It involves digesting the DNA with specific restriction enzymes and applying it to a gel matrix under an electric field that periodically changes direction. PFGE is a modified version of agarose gel electrophoresis that allows for the analysis of bacterial DNA fragments that are significantly larger than those analyzed by conventional restriction enzyme analysis. It provides a comprehensive representation of the entire bacterial chromosome in a single gel, producing distinct and well-resolved DNA fragments with a high level of reproducibility.
Genotyping microorganisms, such as Staphylococcus aureus, is crucial for understanding their global evolution and investigating their genetic relatedness, particularly in epidemiological studies. Various genotyping methods are available for S. aureus, each with its own strengths and weaknesses. These methods include PFGE, surface protein A typing (spa-typing), multi-locus sequence typing (MLST), plasmid profile analysis, and whole-genome DNA sequence typing. S. aureus is a significant cause of life-threatening bacterial infections, both in healthcare settings and the community. It is associated with healthcare-associated infections, surgical site infections, infections involving heart valves and cardiac devices, bacteremia, and endocarditis. Additionally, S. aureus has become increasingly resistant to available antibiotics, with methicillin-resistant S. aureus (MRSA) being a major concern. Therefore, a reliable and highly discriminatory genotyping technique is necessary to differentiate and type these isolates and to better understand the epidemiology and outbreaks of antibiotic-resistant strains.
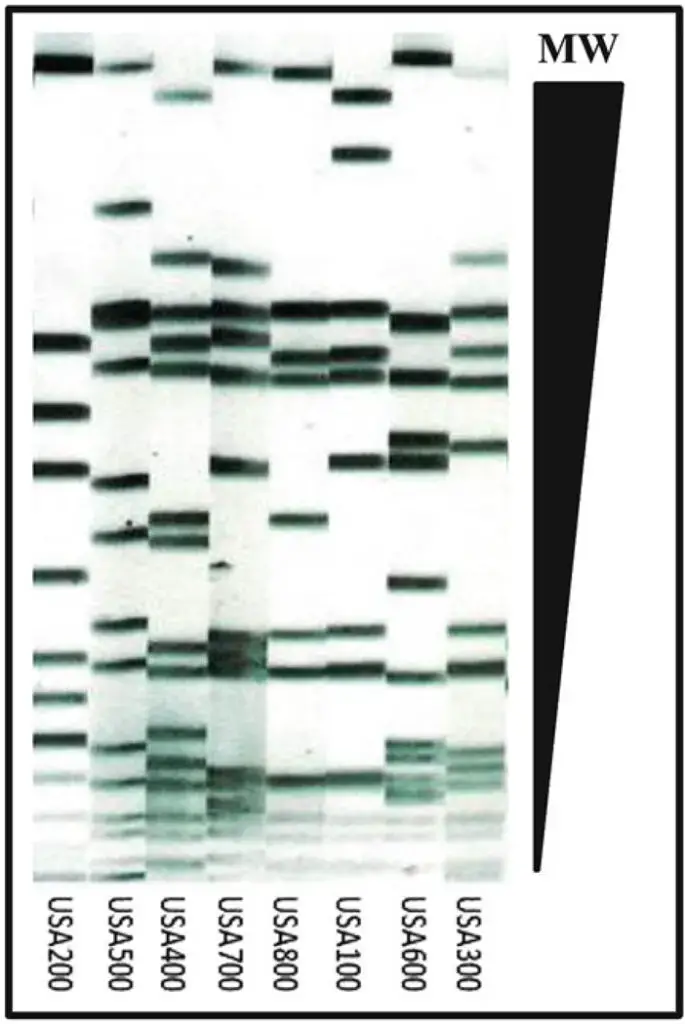
PFGE is often considered the gold standard among the various DNA-based methods for genotyping S. aureus and other bacterial pathogens. It offers high discriminatory power, reproducibility, ease of execution, cost-effectiveness, and wide availability. PFGE involves isolating intact chromosomal DNA by embedding bacterial cells in an agarose plug to prevent DNA shearing. The chromosomal DNA within the agarose plug is then digested with a rare cutting restriction enzyme, resulting in the generation of large DNA fragments. These digested DNA samples, ranging from 10 to 800 kilobases (kb) in size, are separated by applying an alternating electric field between distinct pairs of electrodes. This process allows megabase (Mb) size DNA fragments to migrate at different speeds through the gel pores towards the anode in a size-dependent manner. The reorientation time of DNA fragments is inversely proportional to their size, ensuring good resolution of large DNA fragments in the agarose gel. The obtained gel images are then normalized and analyzed using software, such as BioNumerics, following established criteria for interpreting PFGE patterns. These patterns serve as virtual barcodes that “type” the strains and enable the determination of their relatedness.
In conclusion, PFGE is a powerful genotyping technique used for the separation and analysis of large DNA molecules in bacterial pathogens. Its ability to provide a comprehensive view of the bacterial chromosome and its high discriminatory power make it a valuable tool for understanding the genetic relatedness, epidemiology, and antibiotic resistance profiles of pathogens like S. aureus.
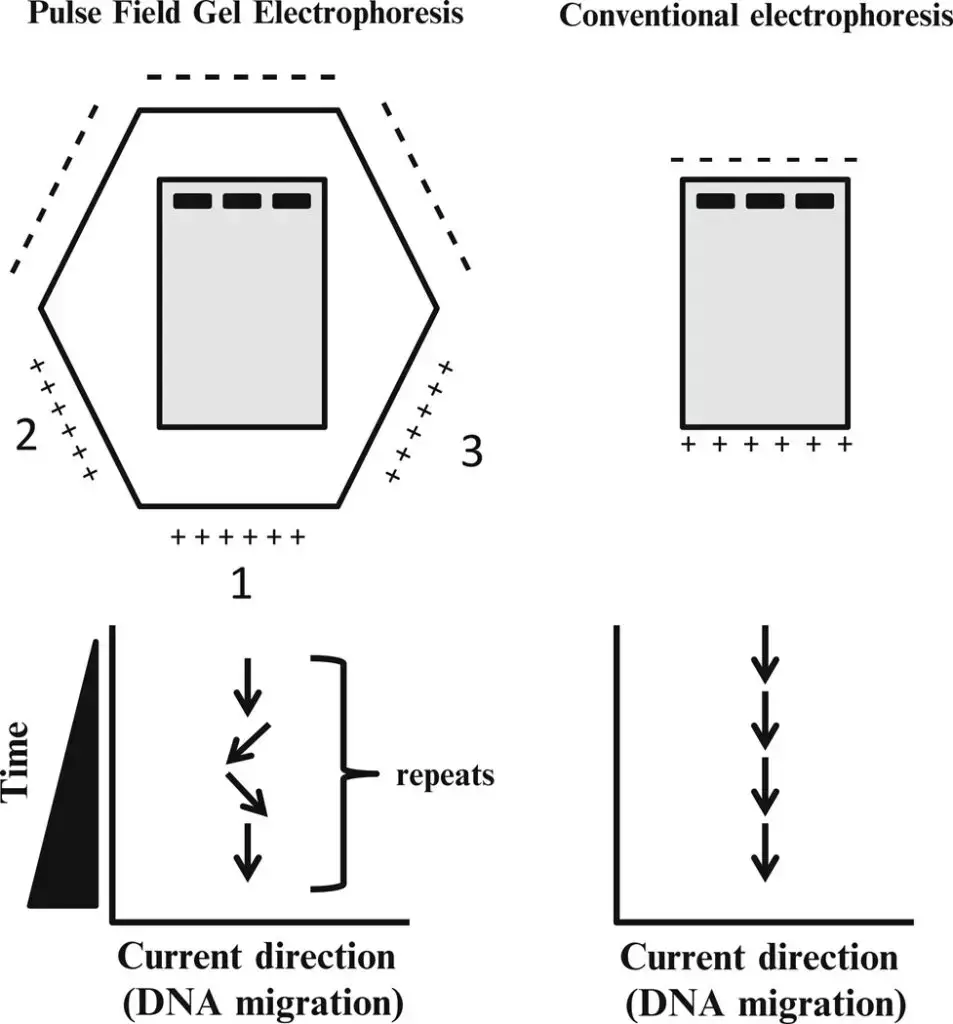
Principle of Pulsed Field Gel Electrophoresis (PFGE)
The principle of Pulsed Field Gel Electrophoresis (PFGE) revolves around the periodic changing of the electric field direction to achieve the separation of large DNA fragments that would otherwise migrate together as a single diffuse band in a gel.
In conventional gel electrophoresis, small DNA fragments move more easily through the gel matrix than larger fragments, resulting in distinct bands according to their size. However, when it comes to very large DNA fragments above a certain threshold length (typically 30-50 kilobases), they tend to migrate at the same rate and appear as a single diffuse band in the gel.
PFGE overcomes this limitation by periodically changing the direction of the electric field. This alternating field causes the different lengths of DNA fragments to react at different rates to the change in field direction. Larger DNA fragments take longer to realign their charge when the field direction is reversed, while smaller fragments realign more quickly. As a result, over time and with the continuous alteration of field direction, each DNA band starts to separate further, even for extremely large DNA fragments.
By introducing periodic changes in the electric field direction, PFGE allows for the separation and resolution of very large DNA pieces that would otherwise be indistinguishable. This principle enables PFGE to be a powerful technique for analyzing and differentiating large DNA molecules, such as entire genomic DNA, and provides researchers with valuable information about the organization and characteristics of the DNA fragments within a sample.
Purpose of Pulse Field Gel Electrophoresis (PFGE)
The purpose of Pulse Field Gel Electrophoresis (PFGE) is to separate large DNA molecules based on their size by applying a pulsed electric field in a gel matrix. PFGE is a powerful molecular biology technique commonly used in genetic and epidemiological studies, particularly in the field of microbial genotyping. It offers higher resolution and discriminatory power compared to traditional gel electrophoresis methods.
The main purposes of PFGE include:
- Genomic DNA Fragmentation: PFGE is used to fragment genomic DNA into large and intact fragments, typically ranging from tens of kilobases to megabases in size. This fragmentation allows for the analysis of the entire genome or specific regions of interest.
- DNA Separation: PFGE separates the DNA fragments based on their size and molecular weight. The pulsed electric field applied during the electrophoresis process causes the DNA fragments to migrate through the gel matrix in a controlled manner, with larger fragments migrating more slowly than smaller ones. This separation enables the visualization and analysis of the DNA fragments.
- Genomic DNA Profiling: PFGE is widely employed in molecular epidemiology and microbial genotyping studies to characterize and compare the genetic profiles of different strains or isolates. By comparing the banding patterns of DNA fragments, PFGE can determine the relatedness or similarity between different isolates, aiding in the investigation of outbreaks, tracking the spread of pathogens, and identifying sources of contamination.
- Subtyping and Strain Differentiation: PFGE is particularly useful in differentiating strains or subtypes within a species. The unique banding patterns obtained from PFGE analysis can be used to identify specific genetic variants or subtypes of microorganisms, providing valuable information for studying genetic diversity, population structure, and evolutionary relationships.
- Outbreak Investigation: PFGE is a critical tool in outbreak investigations, especially for foodborne and nosocomial infections. By comparing the PFGE profiles of pathogens isolated from different patients, food samples, or environmental sources, investigators can establish links between cases, identify the source of contamination, and implement appropriate control measures.
Overall, PFGE is a versatile technique for DNA analysis, offering high resolution and discriminatory power for genotyping and molecular epidemiology studies. Its applications extend to various fields, including microbiology, public health, food safety, and forensic science, where precise DNA characterization and strain differentiation are essential for understanding genetic relationships and investigating disease outbreaks.
Materials Required for Pulse Field Gel Electrophoresis (PFGE)
Pulse Field Gel Electrophoresis (PFGE) is a powerful technique used in molecular biology to separate and analyze large DNA molecules, such as those found in bacteria. The process requires several materials to ensure accurate and efficient results. Here is a list of the materials required for performing PFGE:
- Trypticase Soy Agar (TSA) Plates: These plates are used for growing bacterial cultures. They provide a nutrient-rich medium that supports the growth of bacteria.
- 37 °C Shaking Incubator: This equipment is used to maintain a constant temperature and agitation for bacterial cultures to grow.
- Turbidity Meter or Spectrophotometer: These instruments are used to measure the turbidity or optical density of bacterial cultures. They help determine the bacterial cell concentration, which is essential for preparing cell suspensions.
- Microcentrifuge: A microcentrifuge is used to pellet bacterial cells from suspensions by centrifugation. It allows for the separation of cells from the culture medium.
- Vortex Mixer: A vortex mixer is used to mix solutions and ensure proper homogeneity of reagents.
- Stationary Water Bath: Both 55-60 °C and 37 °C stationary water baths are required for various steps in the PFGE protocol. These baths provide a constant temperature for specific reactions and gel preparation.
- SeaKem Gold Agarose: This high-quality agarose is used to prepare agarose plugs, which encapsulate the bacterial cells for subsequent steps in the PFGE process.
- TE Buffer: TE buffer (10 mM Tris, 1 mM EDTA, pH 8.0) is used as a storage and working solution for various steps in the PFGE protocol. It helps maintain the pH stability and integrity of DNA.
- Microwave Oven: A microwave oven is used to melt agarose and prepare agarose plugs.
- PFGE Plug Mold: This specialized mold, such as the one from Bio-Rad, is used to shape and form agarose plugs containing bacterial cells.
- Lysostaphin Enzyme: Lysostaphin enzyme is used for bacterial cell lysis. It is prepared as a suspension in sodium acetate buffer and helps break down the bacterial cell walls.
- Stainless Steel Spatulas: These spatulas are used to handle and transfer agarose plugs during various steps in the PFGE process.
- EC Lysis Buffer: This buffer contains Tris, EDTA, NaCl, Brij-58, Deoxycholate, and Sarcosyl. It is used for plug lysis and helps solubilize the agarose and release the bacterial DNA.
- Tubes, Pipettes, and Basic Lab Supplies: Various tubes, pipettes (including 5 ml pipettes), and other basic lab supplies are required for handling reagents, plugs, and performing different steps in the PFGE protocol.
- Restriction Enzymes and Buffers: Specific restriction enzymes, such as SmaI and XbaI, are used for digesting the bacterial DNA within the plugs. The enzymes are provided with packaged restriction buffers and bovine serum albumin (BSA) to enhance their activity.
- Cutting Dish, Scalpel, and Bucket of Ice: These items are used to cut the agarose plugs and maintain their integrity during the restriction enzyme digestion process. The cutting dish provides a sterile surface, and the scalpel helps in precise cutting. The bucket of ice or insulated storage box helps keep the samples cool.
- 10× TBE Buffer: TBE buffer (Tris-Borate-EDTA) is used as a running buffer during gel electrophoresis. It provides the appropriate ionic strength and pH for efficient separation of DNA fragments.
- Gel-Casting Platform and Accessories: These include a gel-casting platform, combs, comb holders, and a gel leveling bubble. They are used to create the agarose gel wells and ensure even gel thickness.
- 1.8% SeaKem Gold Agarose Gel: This specific concentration of agarose is used to seal the wells of the gel, preventing DNA leakage during electrophoresis.
- CHEF-DR II System: The CHEF-DR II system from Bio-Rad is a pulsed-field gel electrophoresis apparatus used for running the gels. It provides precise control over the electric field strength and pulse times.
- Ethidium Bromide Solution: Ethidium bromide is a fluorescent dye used for staining DNA in the gel. It intercalates with the DNA molecules and becomes visible under UV light.
- Containers for Staining and Destaining Gels: Covered glass dishes or containers are used for staining and destaining the gels with ethidium bromide solution.
- Gel Documentation Apparatus: A gel documentation apparatus, such as the Gel Doc 2000 from Bio-Rad, is used for visualizing and capturing images of the stained gels. It helps document the results of the PFGE analysis.
- BioNumerics Software: BioNumerics software, specifically version 4.0 from Applied Maths, is used for data analysis and interpretation of the PFGE results. It provides tools for comparing and clustering DNA banding patterns.
These materials are essential for conducting Pulse Field Gel Electrophoresis (PFGE) experiments. It is important to use ultrapure deionized water and high-quality reagents to ensure reliable and reproducible results. Additionally, proper disposal of waste materials should be done following relevant regulations.
Preparation of Bacterial Cells
The preparation of bacterial cells is a crucial step in many molecular biology techniques, including Pulse Field Gel Electrophoresis (PFGE). Here is a step-by-step guide on how to prepare bacterial cells for PFGE, incorporating the mentioned ideas:
- Start by obtaining Trypticase Soy Agar (TSA) plates. These agar plates provide a nutrient-rich medium for bacterial growth. Ensure the plates are sterile before use.
- Inoculate a single colony of the desired bacterial strain onto a TSA plate. Using a sterile inoculation loop or swab, streak the colony onto the surface of the agar plate in a zigzag pattern.
- Incubate the TSA plate in a 37 °C shaking incubator. The shaking motion ensures even distribution of bacterial cells and promotes optimal growth. Incubation times may vary depending on the bacterial strain being cultured. Follow the recommended incubation period for your specific strain.
- After the incubation period, remove the TSA plate from the incubator and visually inspect it. Look for well-isolated colonies that exhibit typical bacterial morphology.
- To prepare a bacterial cell suspension, select a well-isolated colony from the TSA plate using a sterile inoculation loop or swab. Transfer the colony into a suitable container or tube containing an appropriate volume of sterile growth medium, such as sterile broth or saline solution.
- Measure the turbidity or optical density of the bacterial suspension using a turbidity meter or spectrophotometer. This step helps estimate the concentration of bacterial cells in the suspension. Adjust the optical density or turbidity to the desired level based on the experimental requirements.
- If necessary, pellet the bacterial cells from the suspension using a microcentrifuge. Centrifuge the suspension at a suitable speed and duration to collect the cells at the bottom of the tube. Carefully decant or aspirate the supernatant to remove the liquid portion.
- Resuspend the pelleted bacterial cells in an appropriate volume of sterile growth medium or buffer, depending on the subsequent steps in the PFGE protocol. Ensure the resuspended cells are thoroughly mixed using a vortex mixer or by gentle pipetting.
The prepared bacterial cell suspension is now ready for further processing in the PFGE protocol, such as the preparation of agarose plugs or cell lysis. Proper handling and aseptic techniques should be followed throughout the process to maintain the integrity and viability of the bacterial cells.
Preparation of Agarose Plugs
The preparation of agarose plugs is an essential step in the Pulse Field Gel Electrophoresis (PFGE) technique. The following steps outline the process, incorporating the ideas mentioned:
- Begin by setting up a 55-60 °C stationary water bath and a 37 °C stationary water bath. These water baths will be used to control the temperature during different stages of agarose plug preparation.
- Obtain SeaKem Gold agarose, a high-quality agarose commonly used in PFGE. Follow the manufacturer’s instructions for weighing the appropriate amount of agarose required for your experiment.
- Prepare TE buffer, a solution composed of 10 mM Tris and 1 mM EDTA at pH 8.0. To make TE buffer, mix 20 ml of 1 M Tris, pH 8.0, with 4 ml of 0.5 M EDTA, pH 8.0, in a graduated cylinder. Add water to make a final volume of 2,000 ml. Transfer the solution to two 1,000 ml glass screw-top bottles and autoclave. Store the TE buffer at room temperature for up to 6 months.
- Ensure you have a clean beaker or container available for “working” with TE buffer. This container will be used to hold the TE buffer during the preparation of agarose plugs.
- Use a 250 ml screw-capped Erlenmeyer flask to measure the desired volume of TE buffer needed for agarose plug preparation. The volume will depend on the number of plugs required for your experiment.
- In a microwave oven, heat the Erlenmeyer flask containing the TE buffer until the solution reaches a boiling point. Take caution to avoid boiling over or spilling.
- Add the pre-weighed SeaKem Gold agarose to the boiling TE buffer in the Erlenmeyer flask. Stir the mixture gently until the agarose is completely dissolved. Ensure there are no visible agarose particles or clumps.
- Allow the agarose solution to cool down to approximately 50-55 °C, either by leaving it at room temperature or transferring it to the 55-60 °C water bath. The agarose should be hot but not too hot to handle.
- While the agarose solution is cooling, prepare a PFGE plug mold, such as the one from Bio-Rad. Ensure the mold is clean and free of any residues.
- Once the agarose solution has reached the appropriate temperature, carefully pour it into the plug mold, filling each well completely. Take care to avoid introducing any air bubbles into the wells.
- Allow the agarose to solidify at room temperature or by placing the mold in a 4 °C refrigerator for quicker solidification.
- Once the agarose has solidified, remove the solid agarose plugs from the mold using stainless steel spatulas or any suitable tools.
The prepared agarose plugs are now ready for further steps in the PFGE protocol, such as plug lysis and DNA analysis. Store the plugs appropriately to maintain their integrity until further use. Follow proper aseptic techniques and dispose of waste materials according to relevant regulations.
Plug Lysis
Plug lysis is a critical step in Pulse Field Gel Electrophoresis (PFGE) that involves the breakdown of agarose plugs and release of genomic DNA. Here is a step-by-step guide on plug lysis, incorporating the mentioned ideas:
- Set up a 55-60 °C stationary water bath and a 37 °C stationary water bath. These water baths will be used to control the temperature during the plug lysis process.
- Prepare the EC lysis buffer, which is a lysis solution used to break down the agarose plugs and release the DNA. To prepare the EC lysis buffer, follow these steps: a. Mix 5.4 ml of 1 M Tris, pH 8.0; 180 ml of 5 M NaCl; 180 ml of 0.5 M EDTA, pH 8.0; 4.5 g of Brij-58 (Polyoxyethylene 20 Cetyl Ether); 1.8 g of sodium deoxycholate; and 4.5 g of sodium lauroyl sarcosinate in a glass beaker using a magnetic stir bar and low heat. b. Stir the mixture until all the components are completely dissolved. c. Add water to make a final volume of 900 ml. d. Transfer the EC lysis buffer to a screw-top bottle and autoclave it for sterilization. e. Store the EC lysis buffer at room temperature for up to 6 months.
- Prepare tubes to hold the agarose plugs and EC lysis buffer. These tubes should be clean and sterile to avoid contamination.
- Use a spatula to carefully remove the agarose plugs from the mold. Take care not to damage the plugs during the removal process.
- Place the agarose plugs into the tubes prepared for plug lysis. Each tube should contain one agarose plug.
- Add an appropriate volume of EC lysis buffer to each tube containing the agarose plug. The volume of the lysis buffer will depend on the size of the agarose plug and the subsequent steps in the PFGE protocol. Follow the recommended guidelines or experimental protocol.
- Use a 5 ml pipette to mix the agarose plug and EC lysis buffer thoroughly. Pipette up and down gently to ensure the lysis buffer is evenly distributed and fully penetrates the agarose plug.
- After mixing, incubate the tubes containing the agarose plugs and lysis buffer in a 55-60 °C stationary water bath. The incubation time may vary depending on the specific experiment and the bacterial strain being analyzed. Follow the recommended incubation period for plug lysis.
- After incubation, transfer the tubes to a 37 °C stationary water bath for a brief period to equilibrate the temperature.
The agarose plugs have now undergone lysis, releasing the genomic DNA. The lysed plugs can be further processed for DNA extraction, restriction enzyme digestion, or other downstream applications in PFGE analysis. Handle the lysed plugs with care to avoid DNA degradation or contamination.
Plug Washing
After the plug lysis step in Pulse Field Gel Electrophoresis (PFGE), it is necessary to wash the agarose plugs to remove residual lysis buffer and impurities. Here is a step-by-step guide on plug washing, incorporating the mentioned ideas:
- Prepare TE buffer, which is a buffer solution containing 10 mM Tris and 1 mM EDTA at pH 8.0. The preparation of TE buffer was described earlier in the content. Ensure that the TE buffer is available for use during the plug washing step.
- Use a spatula or any suitable tool to hold the agarose plug-containing tube, making sure to handle it gently and avoid any damage to the plug.
- Carefully transfer the agarose plug from the tube containing the lysis buffer to a clean tube using the spatula. Ensure that the plug is securely placed in the new tube without any loss.
- Add an appropriate volume of TE buffer to the tube containing the agarose plug. The volume of TE buffer required for washing will depend on the size of the agarose plug. Follow the recommended guidelines or experimental protocol for the specific volume.
- Use a 5 ml pipette to gently mix the agarose plug and TE buffer. Pipette up and down carefully to ensure thorough mixing and wash the plug effectively.
- Place the tube containing the agarose plug and TE buffer on an orbital rocker, rotator, or any equivalent device at room temperature. The purpose of this step is to allow gentle agitation and facilitate washing by promoting the exchange of buffer around the agarose plug.
- Allow the agarose plug to wash in the TE buffer for a sufficient period. The duration may vary depending on the specific experiment or protocol. Typically, a wash duration of 30 minutes to 1 hour is recommended, but it can be adjusted as needed.
- After the appropriate wash time, remove the tube from the orbital rocker or equivalent device.
- Discard the TE buffer from the tube without losing the agarose plug. Take care not to disrupt or remove the agarose plug during the buffer removal process.
- Repeat the washing process by adding fresh TE buffer to the tube containing the agarose plug. Repeat steps 5 to 9 for the desired number of washes. Multiple washes help to ensure thorough removal of residual lysis buffer and contaminants.
- After the final wash, remove the TE buffer from the tube, ensuring that the agarose plug is retained in the tube.
The agarose plug is now washed and ready for the next step in the PFGE protocol. Handle the washed plugs carefully to avoid any damage or contamination. The washed plugs can be used for DNA digestion, gel casting, and further analysis in PFGE.
Restriction Enzyme Digestion
Restriction enzyme digestion is a crucial step in Pulse Field Gel Electrophoresis (PFGE) that involves the cleavage of genomic DNA by specific restriction enzymes. Here is a step-by-step guide on restriction enzyme digestion, incorporating the mentioned ideas:
- Begin with pretested Salmonella serotype Braenderup strain H9812 plugs. These plugs contain the genomic DNA that needs to be digested by restriction enzymes. Make sure the plugs have been prepared according to the instructions outlined in Section 4.
- Prepare microcentrifuge tubes for the restriction enzyme digestion process. These tubes will hold the agarose plugs and the digestion mixture.
- Use a 5 ml pipette or a standard Pipetman to transfer the agarose plug(s) into the microcentrifuge tube(s). Each tube should contain one agarose plug.
- Prepare the restriction enzyme digestion mixture. This mixture typically includes the restriction enzyme, its appropriate buffer, and any necessary additives such as bovine serum albumin (BSA). Follow the manufacturer’s instructions for the specific restriction enzyme being used.
- Add the appropriate volume of the restriction enzyme buffer to the microcentrifuge tube(s) containing the agarose plug(s). The volume of the buffer will depend on the experimental protocol and the recommended enzyme-to-buffer ratio.
- Add the restriction enzyme to the microcentrifuge tube(s), following the recommended amount specified by the manufacturer. It is important to use the correct amount of enzyme for efficient digestion.
- If necessary, add bovine serum albumin (BSA) to the mixture according to the manufacturer’s instructions. BSA is often added to enhance enzyme activity and stability.
- Use sterile, Type I water to adjust the final volume of the digestion mixture in each microcentrifuge tube, if needed. Ensure that the volume is appropriate for efficient digestion and mixing.
- Gently mix the contents of the microcentrifuge tube(s) using a pipette or by inverting the tubes several times. Ensure that the enzyme, buffer, and water are thoroughly mixed with the agarose plug(s).
- Place the microcentrifuge tube(s) in a cutting dish, which can be a sterile disposable petri dish or any equivalent sterile container. This dish will hold the tubes during the subsequent steps.
- Use a sharp scalpel or razor blade to carefully cut the agarose plug(s) into smaller pieces within the microcentrifuge tube(s). This step increases the surface area of the plugs, allowing better contact between the restriction enzyme and the DNA.
- After cutting the plugs, place the cutting dish containing the microcentrifuge tube(s) on a bucket of ice or in a −20 °C insulated storage box. Cooling the samples helps to slow down the reaction and prevents DNA degradation.
- Incubate the microcentrifuge tube(s) in the cutting dish at the appropriate temperature for the specific restriction enzyme being used. Follow the manufacturer’s instructions for the recommended incubation temperature and duration.
- During the incubation period, gently rock the cutting dish on an orbital rocker, rotator, or any equivalent device at room temperature. This promotes proper mixing and ensures even distribution of the digestion mixture over the agarose plug(s).
- After the incubation time, remove the microcentrifuge tube(s) from the cutting dish and stop the digestion reaction according to the manufacturer’s instructions. This is typically done by heating the samples or adding specific reagents to deactivate the enzyme.
The agarose plugs have now undergone restriction enzyme digestion, resulting in the cleavage of the genomic DNA. The digested DNA fragments can be analyzed further using gel electrophoresis or processed for downstream applications in PFGE. Handle the digested plugs and samples with care to avoid DNA degradation or contamination.
Preparing and Running the Gel
Preparing and running the gel is a critical step in Pulse Field Gel Electrophoresis (PFGE) for separating DNA fragments based on their size. Here is a step-by-step guide incorporating the mentioned ideas:
- Prepare a 10× TBE buffer, which serves as the running buffer for the gel. TBE buffer consists of Tris base, boric acid, and EDTA. Ensure that the buffer is properly prepared according to the specific protocol or commercial product instructions.
- Set up a stationary water bath at a temperature between 55–60 °C. This will be used to melt the agarose and maintain it in a liquid state during gel preparation.
- Obtain sterile distilled water and pre-warm it to 55 °C. This water will be used to prepare the agarose gel.
- Use SeaKem Gold agarose (Bio-Rad #161–3109) or any other appropriate agarose product for gel preparation. Weigh the desired amount of agarose according to the gel size and concentration required for your experiment.
- In a heat-resistant container, combine the weighed agarose with the pre-warmed distilled water. The exact ratio of agarose to water will depend on the desired gel concentration. Stir the mixture thoroughly to ensure the agarose is completely dissolved.
- Add the appropriate volume of the 10× TBE buffer to the agarose mixture. The volume of TBE buffer required will depend on the specific protocol or gel concentration being used. Stir the mixture gently to ensure uniform distribution of the buffer.
- Place the agarose mixture in the stationary water bath at 55–60 °C. Allow the agarose to melt completely, periodically swirling or gently stirring to facilitate the process. Avoid excessive agitation that could introduce air bubbles.
- While the agarose is melting, prepare the gel-casting platform and accessories. This includes assembling the gel tray, comb, and comb holder according to the specific equipment instructions.
- Once the agarose has melted, remove it from the water bath and allow it to cool slightly until it reaches a temperature suitable for casting the gel. Ensure that the agarose is still in a liquid state but not too hot to handle.
- Pour the melted agarose into the gel tray, ensuring that it is level and fills the entire tray to the desired height. Take care to avoid introducing air bubbles during pouring.
- Insert the appropriate comb into the comb holder, ensuring that it is properly aligned with the gel tray. The comb creates wells in the gel where the DNA samples will be loaded.
- Allow the agarose gel to solidify completely at room temperature. This typically takes around 30 minutes to 1 hour, but the exact time will vary depending on the gel size and concentration.
- Once the gel has solidified, carefully remove the comb by gently pulling it straight upward. This will leave wells in the gel for sample loading.
- Prepare a 1.8% SeaKem Gold agarose gel or any other appropriate concentration to seal the wells at the ends of the gel. This prevents sample leakage during electrophoresis.
- Place the gel-casting platform containing the solidified gel into the CHEF-DR II system or any other suitable electrophoresis apparatus. Follow the manufacturer’s instructions for proper installation and setup.
- Fill the electrophoresis chamber with the 10× TBE buffer until it covers the gel completely. This buffer will serve as the medium for the electrical current during gel electrophoresis.
- Load the DNA samples into the wells of the gel using a pipette or other suitable loading method. Be careful not to overload the wells or introduce air bubbles.
- Attach the lid to the electrophoresis chamber, ensuring that the electrodes are properly connected to the power supply.
- Set the appropriate running conditions on the CHEF-DR II system or your chosen apparatus, including voltage, run time, and pulse duration. These parameters will depend on the size range of the DNA fragments you are analyzing.
- Start the gel electrophoresis run, and monitor the progress as the DNA fragments migrate through the gel under the influence of the electric field. Follow the recommended running time based on the expected separation of DNA fragments.
- After the electrophoresis run is complete, carefully remove the gel from the apparatus. Handle the gel with clean, gloved hands to avoid contamination.
- The gel can be visualized using various staining methods, such as ethidium bromide staining or fluorescent dyes, depending on the specific experimental requirements.
Remember to follow appropriate safety protocols, dispose of waste materials properly, and consult the specific instructions provided by the manufacturer or protocol you are following.
Staining and Documentation
Staining and documentation are crucial steps in Pulse Field Gel Electrophoresis (PFGE) to visualize and analyze DNA fragments separated in the gel. Here is a guide incorporating the mentioned ideas:
- Prepare an ethidium bromide solution with a concentration of 10 mg/ml. This can be obtained from a commercial supplier, such as AMRESCO #X328, or an equivalent product. Follow the instructions provided by the manufacturer for proper handling and dilution of the ethidium bromide solution.
- Obtain containers, preferably covered glass dishes, to hold the gel during staining and destaining processes. These containers should be of an appropriate size to accommodate the gel comfortably.
- Fill the containers with distilled water, approximately 2 liters, to create a bath for rinsing and washing the gel. The distilled water should be clean and free from contaminants.
- Place the gel into one of the containers filled with distilled water. Gently agitate the container to rinse the gel and remove any excess buffer or debris from the gel surface.
- Transfer the gel to a fresh container filled with the ethidium bromide solution. Ensure that the gel is fully immersed in the staining solution and allow it to stain for the recommended time according to the specific protocol or manufacturer’s instructions. This time can vary depending on the concentration of ethidium bromide and the desired level of staining.
- After the staining period, carefully remove the gel from the ethidium bromide solution and place it into another container filled with distilled water. Gently agitate the container to rinse off excess staining solution from the gel.
- Repeat the rinsing process by transferring the gel to a second container of distilled water. This step is crucial to remove any remaining ethidium bromide that may interfere with subsequent visualization or analysis.
- Prepare the gel documentation apparatus, such as the Gel Doc 2000 from Bio-Rad or an equivalent system, according to the manufacturer’s instructions. This typically involves setting up the appropriate filters, camera settings, and lighting conditions for gel imaging.
- Place the gel onto the gel documentation apparatus, ensuring that it is properly positioned for optimal imaging. Follow the specific instructions provided by the manufacturer to capture high-quality gel images.
- Document the gel images using the gel documentation apparatus. This may involve capturing images under both UV and visible light, depending on the staining method and detection system used. Follow the guidelines provided by the manufacturer for proper image acquisition and storage.
- Analyze the gel images using suitable software or analysis tools. This may involve quantifying band sizes, comparing patterns, or performing molecular weight estimations. Commercial software such as BioNumerics can be used for data analysis.
Ensure that appropriate safety measures are followed when handling ethidium bromide and other staining reagents. Properly dispose of waste materials according to local regulations.
Protocol of Pulse Field Gel Electrophoresis (PFGE)
Overview
- Day 0: Streak plates.
- Day 1: Make plugs.
- Day 2: Wash plugs.
- Day 3: Restriction digest and electrophoresis.
- Day 4: Stain, take photograph, analyze in Bionumerics.
Day 0: Streak plates
Streak plates are a common method used in microbiology to isolate and culture bacterial colonies. Here’s a guide based on the provided content:
- Obtain sterile Trypticase Soy Agar (TSA) plates. These plates provide a nutrient-rich medium that supports the growth of various microorganisms.
- Using a sterile inoculating loop or a sterile bacteriological loop, obtain a sample of Staphylococcus aureus culture or isolate. Ensure that the loop is properly sterilized by passing it through a flame until it becomes red-hot. Allow the loop to cool briefly before proceeding.
- Lift the lid of a TSA plate just enough to access a small area on the surface of the agar. This area will be used for streaking the bacterial sample.
- Dip the cooled sterilized loop into the Staphylococcus aureus culture or isolate, taking care to obtain a small amount of the bacterial sample. The sample should be evenly distributed across the loop.
- Carefully streak the bacterial sample onto the TSA plate by lightly dragging the loop back and forth across the surface of the agar in a pattern that resembles an elongated “S” shape. Start from the top of the agar and progressively move towards the bottom. This streaking technique helps to dilute the bacterial sample and separate individual cells.
- After streaking the initial sample, sterilize the loop again by passing it through a flame. Allow it to cool briefly.
- Rotate the TSA plate approximately 90 degrees, ensuring that the previously streaked area is facing away from you.
- Using the cooled, sterilized loop, pick up a small amount of bacteria from the streaked area and drag it across the agar in a new section adjacent to the first streak. This second streaking helps to further dilute the bacteria and promote the growth of isolated colonies.
- Repeat the sterilization and streaking process two more times, rotating the plate 90 degrees after each streak. With each streak, transfer fewer bacteria to the plate, allowing for the isolation of individual colonies.
- Once the streaking is complete, close the lid of the TSA plate securely and label it with the necessary information, such as the date, sample name, and any other relevant identifiers.
- Incubate the streak plate upside down at 37 °C in an incubator for 18-24 hours. This temperature and duration provide optimal conditions for the growth of Staphylococcus aureus.
- After the incubation period, observe the streak plate for the presence of isolated colonies. Each isolated colony represents a single bacterial cell that has multiplied into a visible cluster.
Streak plates allow for the isolation of individual bacterial colonies, which can be further characterized or subjected to additional testing. Proper aseptic techniques should be followed during the entire process to avoid contamination. Dispose of used agar plates and sterilized loops according to appropriate laboratory protocols and guidelines.
Day 1: Make plugs
To make plugs for further analysis or experiments, follow these steps based on the provided content:
- Begin by preparing the necessary equipment and reagents. Turn on 37 °C and 55 °C water baths and prepare two boxes of ice for later use.
- Label a 15 ml conical tube, a 5 ml polystyrene round-bottom tube, and a 1.5 ml microcentrifuge tube for each sample you will be working with.
- Add 2 ml of sterile water to each 5 ml polystyrene round-bottom tube. This will serve as the initial suspension medium.
- Add 3 ml of EC lysis buffer to each 15 ml conical tube. The EC lysis buffer helps in breaking down the cells and releasing their genetic material.
- Using a sterile swab, collect cells from the desired source (e.g., a plate) and suspend them in water in the 5 ml polystyrene round-bottom tube. Vortex the tube briefly to ensure proper mixing. Place the tube in a turbidity meter and aim for a reading between 0.80 and 0.89. Adjust the suspension by adding more sterile water in increments of 1.0 ml if the reading is too high or more cells if the reading is too low. Store the tubes on ice.
- Transfer 200 µl of the suspended cells to a 1.5 ml microcentrifuge tube. Centrifuge the tubes at 13,000 rpm for 6 minutes. This step helps to pelletize the cells.
- While the samples are being centrifuged, prepare the agarose for gram-positive bacteria. Combine 0.9 g of SeaKem Gold agarose with 50 ml of TE buffer in a 200 ml screw-top flask. Microwave the flask for 1 minute and 50 seconds, then gradually loosen the lid and swirl the agarose. If it is not completely melted, continue microwaving in 25-second intervals, loosening the lid and swirling after each time, until the agarose is thoroughly melted. Place the flask in the 55 °C water bath to equilibrate for at least 30 minutes.
- Once the centrifugation is complete, use a sterile pipette to aspirate the entire supernatant from the microcentrifuge tubes and discard it. The pellet should now be visible and measure around 2-3 mm in diameter.
- Add 300 µl of TE buffer to each microcentrifuge tube containing the cell pellet and vortex to resuspend the cells. Place the tubes in a 37 °C water bath for 10 minutes to allow the cells to recover.
- For each isolate, label two wells on the plug mold. Remove the microcentrifuge tubes from the 37 °C water bath.
- Working with one tube at a time, add 3 µl of Lysostaphin (1 mg/ml) and 300 µl of the 55 °C agarose (prepared in step 7) to the microcentrifuge tube. Mix quickly but gently by pipetting the mixture ten times. Using a sterile pipette, fill two wells of the plug mold, slightly overfilling to produce a rounded top.
- Repeat step 11 for each sample, ensuring that you fill the appropriate wells on the plug mold. Allow the plugs to harden for 10-15 minutes at room temperature or for 5 minutes in an 8 °C refrigerator.
- Once the plugs have hardened, use the provided snap-off tool or a spatula cleaned with ethanol to push the plugs (two per sample) into the 15 ml conical tubes containing approximately 3 ml of EC lysis buffer. Ensure that the plugs are fully immersed in the buffer.
- Incubate the tubes in a 37 °C water bath for at least 4 hours, preferably overnight. This step allows the lysis buffer to act on the cells within the plugs, breaking down their membranes and releasing the genetic material of interest.
Following these steps will enable you to create plugs containing the desired bacterial material for further analysis, such as DNA extraction or other molecular techniques.
Day 2: Wash plugs
To wash the plugs, follow these steps based on the provided content:
- Pour off the EC lysis buffer from the conical tubes into a glass beaker, making sure to hold the cap close to the rim to prevent any plugs from escaping. Check the beaker for any plugs that may have accidentally escaped during the transfer.
- Add 5 ml of TE buffer (Tris-EDTA buffer) to each tube, ensuring that all plugs are fully immersed in the buffer. TE buffer is commonly used in molecular biology applications for its ability to stabilize DNA and RNA.
- Securely cap the tubes and place them horizontally in a glass tray on a rocker table. Set the speed of the rocker table to approximately 60 rpm. This gentle rocking motion helps in the washing process by allowing the buffer to circulate around the plugs.
- Allow the plugs to wash for 60 minutes at room temperature. The rocking motion ensures thorough washing of the plugs.
- After the washing time is complete, carefully pour off the buffer from the tubes into the glass beaker, following the same precautions as in step 1.
- Repeat the wash two more times, adding fresh TE buffer each time and washing for 60 minutes. This will make a total of three washes, ensuring effective removal of residual lysis buffer and debris.
- If desired, the last wash can be done overnight. Simply leave the tubes on the rocker table with fresh TE buffer and resume the process the next day.
- Once the washes are complete, the plugs can be stored refrigerated until all reagents are prepared for the subsequent enzyme digestion step.
By following these steps, you can effectively wash the plugs and remove any remaining lysis buffer, preparing them for the next stage of the experimental procedure.
Day 3: Restriction digest and electrophoresis
To perform the restriction digest and electrophoresis, follow these steps based on the provided content:
- Turn on the 37 and 55 °C water baths and prepare a box of ice. Remove the 10× multicore stock, SmaI, XbaI, and BSA from the freezer and allow them to thaw at room temperature. Centrifuge the reagents for 1 minute and then place them on ice.
- Label a 1.5 ml microcentrifuge tube for each sample, including a staphylococcal control (NCTC 8325) and four Salmonella size standards.
- In a 15 ml conical tube, prepare the restriction buffer by combining 420 µl of the 10× multicore stock with 3,780 µl of sterile water. This quantity is sufficient for 21 samples. Adjust the volumes accordingly based on the number of samples.
- Add 200 µl of the prepared restriction buffer to each labeled microcentrifuge tube.
- Clean a petri dish, spatula, and scalpel with ethanol. Place a plug in the petri dish and use the scalpel and spatula to cut two slices measuring 2–3 mm in thickness (five slices for Salmonella samples). Transfer these slices to the corresponding microcentrifuge tube, and return the remaining portion of the plug to its original tube. Repeat this step for each sample, ensuring to clean the dish, spatula, and scalpel each time. Store the slices and remaining plugs at 8 °C.
- Allow the staphylococcal samples to equilibrate at room temperature for 30–45 minutes, and place the Salmonella samples (prepared separately) in a 37 °C water bath for the same duration.
- While keeping all reagents and preparation tubes on ice at all times, prepare the restriction enzyme mix for the staphylococcal samples by combining 360 µl of the 10× multicore stock, 54 µl of SmaI (10 U/µl), 36 µl of acetylated BSA, and 3,150 µl of sterile water in a 15 ml conical tube. Invert the tube, vortex it, and then return it to ice. Adjust the volumes accordingly based on the number of samples.
- While keeping all reagents and preparation tubes on ice at all times, prepare the restriction enzyme mix for the Salmonella samples by combining 100 µl of the 10× multicore stock, 20 µl of XbaI (10 U/µl), 10 µl of acetylated BSA, and 869 µl of sterile water in a 1.5 ml microcentrifuge tube. Invert the tube, vortex it, and then return it to ice. Adjust the volumes accordingly based on the number of samples.
- Remove the buffer from the plugs using disposable SAMCO pipettes, ensuring the plugs are clear of any remaining lysis buffer.
- Add 200 µl of the appropriate restriction enzyme/buffer mix (SmaI for staphylococcal samples or XbaI for Salmonella samples) to each microcentrifuge tube containing the plugs.
- Incubate the tubes for 3–4 hours at room temperature for staphylococcal samples, or in a 37 °C water bath for Salmonella samples. This incubation allows the restriction enzymes to cleave the DNA at specific recognition sites.
By following these steps, you can perform the restriction digest and prepare the samples for subsequent electrophoresis, which will help analyze and visualize the DNA fragments obtained after digestion.
Day 4: Stain, take photograph, analyze in Bionumerics
1. Stain Gel
To stain the gel, follow these steps based on the provided content:
- Add 30 µl of 1 mg/ml ethidium bromide to 300 ml of purified water in a glass tray. Adjust the volume accordingly based on the size of the tray you are using. Ethidium bromide is a fluorescent dye commonly used to visualize DNA in the gel.
- Carefully slide the gel off its black backing and place it into the tray containing the ethidium bromide solution. Ensure that the gel is fully submerged in the staining solution.
- Incubate the gel at room temperature for 45 minutes. This allows the ethidium bromide to bind to the DNA within the gel, making it visible under ultraviolet (UV) light.
- After the incubation period, carefully remove the gel from the staining solution and transfer it to a clean container or tray.
- To decolorize the gel, immerse it in 1 liter of purified water. The decolorization process helps remove excess ethidium bromide from the gel.
- Allow the gel to decolorize in the water for 90 minutes. During this time, the ethidium bromide will diffuse out of the gel.
Once the staining and decolorization steps are complete, the gel is ready for further analysis or documentation. Please note that ethidium bromide is a hazardous substance, and appropriate safety precautions should be taken when handling and disposing of it.
2. Photograph Gel (Picture Can Be Captured in Any Equivalent Way)
To photograph the gel and capture an image, you can follow these steps based on the provided content:
- Carefully slide the gel back onto the black backing to remove it from the water and prepare it for imaging. Ensure that the gel is securely positioned on the backing.
- Transfer the gel to the UV transilluminator, which is a device that emits UV light to visualize the fluorescently stained DNA in the gel.
- Slide the gel off the black backing and place it on the transilluminator table. Ensure that the gel is properly aligned and centered on the table.
- Position the camera and a hood over the table. The hood helps to block external light and minimize background interference in the image.
- Turn on the UV light on the transilluminator. The UV light will cause the ethidium bromide-stained DNA bands in the gel to fluoresce, making them visible for imaging.
- Set the camera to the appropriate settings. Set the image type to RAW, which allows for higher quality and more flexibility during post-processing. Adjust the exposure time to 15 seconds to capture a properly exposed image.
- Zoom in on the gel to frame the desired area for imaging. Ensure that the gel bands are clearly visible and properly aligned within the frame.
- Take at least two pictures of the gel, ensuring that the gel is in focus and properly exposed in each shot. This can help ensure that you have multiple images to choose from and reduce the risk of capturing a blurry or improperly exposed image.
Once the images are captured, they can be transferred to a computer or other image processing software for analysis, documentation, and further manipulation if needed.
3. Processing PFGE Images Using BioNumerics
To process PFGE (Pulsed-Field Gel Electrophoresis) images using BioNumerics software, you can follow these steps based on the provided content:
- Open the BioNumerics software and click on “Add new experiment file” to import the .TIF image file of the PFGE gel. This will load the gel image into the software for further processing and analysis.
- Once the gel image is loaded, follow the instructions provided by the software to process the TIFF image. This typically involves a series of steps to enhance the image quality and extract relevant information from the gel.a. Convert the TIFF image to gel strips: The software will provide tools to convert the gel image into individual gel strips, separating each DNA band lane for further analysis.b. Define curves: Define the curves or lanes for each gel strip, accurately marking the positions of the DNA bands. This step helps the software identify and analyze the bands correctly.c. Normalize the gel: Normalize the gel image by adjusting brightness, contrast, and other parameters to standardize the image quality and remove any variations caused by the gel staining or imaging conditions.d. Find gel bands: The software will use pattern recognition algorithms to automatically detect and identify the DNA bands in each gel strip. This step is crucial for subsequent analysis and comparison of the PFGE patterns.
- After the image processing steps, you can proceed with cluster analysis to compare the isolates. In the software, select the isolates or samples that you want to compare and click on “Calculate Cluster Analysis” or a similar option. The software will guide you through the specific instructions for performing cluster analysis.Cluster analysis helps to group similar PFGE patterns together, allowing you to identify related isolates or strains based on their DNA banding patterns. This analysis can be useful for studying genetic relatedness, identifying outbreaks, or understanding the population structure of microorganisms.
Preparation of Salmonella PFGE Plugs
- The processing of PFGE (Pulsed-Field Gel Electrophoresis) images using BioNumerics software involves several steps, including the preparation of Salmonella plugs as standards, casting the plugs, lysing the cells in agarose plugs, and washing the plugs. These steps ensure the accurate analysis and comparison of DNA fragments in the PFGE gels.
- To begin with, Salmonella plugs should be used as standards in each gel to ensure consistency and provide a reference point for analysis. The standard strain used is Salmonella serotype Braenderup strain H9812. On Day 0, the strain is streaked onto TSA plates and incubated at 37 °C for 18-24 hours.
- On Day 1, the plugs are prepared. The necessary equipment and reagents should be prepared beforehand. A 1% SeaKim Gold agarose gel is prepared by mixing 0.5 g of SeaKim Gold agarose with 47.5 ml of TE buffer. The mixture is dissolved and equilibrated in a 55 °C water bath. Then, 2.5 ml of 20% SDS (sodium dodecyl sulfate) is added to the agarose gel mixture, and it is kept in the water bath until ready to use.
- A Cell Suspension Buffer is prepared using Tris and EDTA, and it is stored in a screw-top bottle. This buffer is used to suspend the cells collected from the TSA plates. A turbidity meter is used to adjust the cell suspension to a desired reading of 0.48-0.52. The buffer with cells is kept on ice.
- Next, the casting of plugs is performed. PFGE plug molds are labeled, and 400 µl of the cell suspension with the strain in the Cell Suspension Buffer is transferred to labeled 1.5 ml microcentrifuge tubes. Then, 20 µl of Proteinase K is added to each tube, followed by the addition of 400 µl of the melted 1% SDS agarose. The mixture is gently mixed and immediately dispensed into the appropriate wells of the plug molds. Care should be taken to avoid bubble formation. The plugs are allowed to solidify at room temperature or can be placed in the refrigerator for a short time.
- After casting the plugs, the cells in the agarose plugs need to be lysed. Labeled polypropylene screw-cap tubes are prepared, and a Cell Lysis Buffer is made using Tris, EDTA, and Sarcosyl. The buffer is heated and dissolved, and then a Cell lysis buffer/Proteinase K mix is prepared by adding Proteinase K to the Cell Lysis Buffer. The plugs are pushed out into the tubes, ensuring they are fully immersed in the buffer. The tubes are incubated in a shaker incubator at 54 °C with vigorous shaking for 1.5-2 hours.
- After cell lysis, the plugs are washed to remove contaminants. Sterile purified water is preheated and added to the tubes for washing the plugs. The tubes are shaken vigorously in a shaker incubator at 50 °C, and the water is poured off. The wash process is repeated once more. Then, preheated sterile TE buffer is added to the tubes for further washing, and the process is repeated four times.
- Finally, the plugs are stored in microcentrifuge tubes with TE buffer at 4 °C until they are ready to be used for PFGE analysis.
- The processed PFGE images can then be loaded into BioNumerics software for further analysis. The software allows for the comparison of DNA banding patterns and the creation of dendrograms to determine the relatedness between different isolates. This analysis is crucial for epidemiological investigations and understanding the genetic diversity and relatedness of bacterial strains.
- It is important to follow all the steps carefully, including the preparation of reagents using ultrapure deionized water and analytical grade reagents. Additionally, proper waste disposal regulations should be followed when disposing of waste materials generated during the process.
- Processing PFGE images using BioNumerics software and following the recommended protocols for plug preparation and analysis allows for accurate and reliable characterization of bacterial strains, aiding in surveillance, outbreak investigations, and other molecular epidemiology studies.
Make Plugs
To make plugs for PFGE analysis, the following steps can be followed:
- Begin by turning on the 54 °C shaker incubator and the water bath set at 55 °C. Prepare ice in a Styrofoam box to keep certain reagents and samples cold during the process.
- Prepare the 1% SeaKim Gold agarose gel for the Salmonella plugs. In a 200 ml screw-top flask, mix 0.5 g of SeaKim Gold agarose with 47.5 ml of TE buffer. Dissolve the mixture by following the instructions in Section 3.3, Step 8. Once dissolved, place the flask in the 55 °C water bath for about 5 minutes to equilibrate the temperature.
- Preheat 2.5 ml of 20% SDS solution to 55 °C. After the agarose gel has equilibrated, add the preheated SDS solution to the gel mixture. Mix the solution thoroughly and keep it in the 55 °C water bath until it is ready to be used.
- Prepare the Cell Suspension Buffer, which is a solution used to suspend the bacterial cells. To do this, mix 10 ml of 1 M Tris (pH 8.0) with 20 ml of 0.5 M EDTA (pH 8.0) in a graduated cylinder. Add water to make a final volume of 100 ml. Transfer the solution to a screw-top bottle and autoclave it for sterilization. The Cell Suspension Buffer can be stored at room temperature for up to 6 months.
- Transfer 2 ml of the Cell Suspension Buffer to labeled 5 ml polystyrene round-bottom tubes. Keep the tubes with the buffer on ice to maintain their cold temperature.
- Using a sterile swab, collect cells from the TSA plate that contains the desired Salmonella strain. Suspend the collected cells in the Cell Suspension Buffer by vortexing the tube briefly to ensure even distribution. Place the tube containing the cell suspension in a turbidity meter. The aim is to achieve a reading of 0.48-0.52 on the turbidity meter, indicating an appropriate cell density. If the reading is too high, add more buffer in 500 µl increments. Conversely, if the reading is too low, add more cells. Store the tubes with the cell suspension on ice until they are ready to be used for plug preparation.
These steps ensure the proper preparation of the Salmonella plugs, which will be further processed for PFGE analysis. The plugs are essential for loading the bacterial DNA into the gel and performing subsequent DNA fragment separation to generate the banding patterns used for strain comparison and analysis.
Casting Plugs
To cast plugs for PFGE analysis, the following steps can be followed:
- Begin by labeling the wells of the PFGE plug molds. The number of plug molds needed can vary, but typically 40 plug molds or even as few as 20 plug molds are used.
- Transfer 400 µl of the cell suspension containing the desired strain in the Cell Suspension Buffer to labeled 1.5 ml microcentrifuge tubes. Ensure that you have more than 15 tubes to accommodate the number of samples.
- Add 20 µl of Proteinase K (20 mg/ml) to each microcentrifuge tube containing the cell suspension. Gently mix the contents of each tube with a pipette tip, working on one or two tubes at a time. Ensure thorough mixing to evenly distribute the Proteinase K.
- Add 400 µl of the previously melted 1% SDS agarose to each microcentrifuge tube containing the cell suspension. The agarose should be brought to room temperature before adding it to the tubes. If the cell suspensions are cold, you can place the tubes in a 37 °C water bath for a few minutes to warm them.
- Mix the agarose and cell suspension gently using a fine-tip transfer pipette. Ensure proper mixing to create a homogenous mixture without introducing any air bubbles.
- Fill the plug molds immediately by dispensing the agarose mixture into the appropriate well(s) of the reusable plug mold. Take care to avoid the formation of bubbles while filling the wells. The amount of cell suspension and agarose provided allows for the creation of two plugs per sample.
- Allow the plugs to solidify at room temperature for approximately 10-15 minutes. Alternatively, if desired, you can place the plug molds in the refrigerator at 4 °C for 5 minutes to expedite the solidification process.
These steps ensure the proper casting of plugs, which are essential for PFGE analysis. The plugs contain the bacterial cells embedded in the agarose matrix and serve as the starting point for subsequent steps in the PFGE procedure, including cell lysis and DNA digestion. The solidified plugs will be further processed to extract the bacterial DNA and perform DNA fragment separation using PFGE.
Lysis of Cells in Agarose Plugs
To perform the lysis of cells in agarose plugs for PFGE analysis, follow these steps:
- Begin by labeling 50 ml polypropylene screw-cap tubes with culture numbers. You will need ten tubes for this step.
- Prepare the Cell Lysis Buffer. Mix 25 ml of 1 M Tris, pH 8.0, with 50 ml of 0.5 M EDTA, pH 8.0, and 5 g of Sodium Lauroyl Sarcosinate in a graduated cylinder. Add water to make a final volume of 500 ml. Transfer the mixture to a 1 l screw-top bottle and warm it to 50-60 °C for 30-60 minutes. Alternatively, you can leave it at room temperature for 2 hours to completely dissolve Sodium Lauroyl Sarcosinate. Autoclave the Cell Lysis Buffer for 20 minutes and store it at room temperature for up to 6 months.
- Prepare the Cell lysis buffer/Proteinase K mix. Add 250 µl of Proteinase K solution (20 mg/ml) to 50 ml of the Cell Lysis Buffer and mix well. This mixture will help break down the proteins and facilitate cell lysis.
- Add 5 ml of the Cell lysis buffer/Proteinase K mix to each of the labeled tubes. These tubes should be the ten polypropylene screw-cap tubes prepared earlier.
- Push out 3-4 agarose plugs into each tube, ensuring that the plugs are fully immersed in the Cell lysis buffer. The agarose plugs contain the bacterial cells and are ready for lysis.
- Place the tubes in a rack and incubate them in a 54 °C shaker incubator. Set the incubator to shake vigorously at a speed of 150-175 rpm. Allow the tubes to incubate for 1.5-2 hours. The incubation period, along with the shaking, helps in the efficient lysis of the bacterial cells within the agarose plugs.
During this step, the Cell Lysis Buffer, along with Proteinase K, breaks down the bacterial cell walls and releases the DNA. The incubation at an elevated temperature enhances the enzymatic activity of Proteinase K, aiding in the effective lysis of the cells. The resulting lysate will be used for further DNA processing and analysis in the PFGE workflow.
Washing of Agarose Plugs
To wash the agarose plugs after lysis of cells in the PFGE (Pulsed-Field Gel Electrophoresis) process, follow these steps:
- Preheat sterile purified water to 50 °C. You will need approximately 10 tubes with 10-15 ml of water each, totaling 200-300 ml. This preheated water will be used to wash the plugs.
- Remove the tubes containing the plugs from the shaker incubator and carefully pour off the Cell lysis buffer/Proteinase K solution. It is important to remove the buffer completely before proceeding with the washing steps.
- Add 10-15 ml of the preheated sterile purified water to each tube containing the plugs. Ensure that each tube is properly capped and tightly sealed. Shake the tubes vigorously at a speed of 150-175 rpm in the 50 °C shaker incubator. Allow the plugs to be washed in the water for 10-15 minutes.
- After the designated time, carefully pour off the water from the plugs. It is essential to remove all the water without losing the plugs. Repeat the wash process one more time by adding another 10-15 ml of preheated sterile purified water to each tube. Shake the tubes vigorously again for 10-15 minutes and pour off the water.
- While the plugs are being washed with water, preheat sterile TE (Tris-EDTA) buffer to 50 °C. You will need approximately 10 tubes with 10-15 ml of TE buffer each, totaling 400-600 ml. TE buffer is commonly used for DNA storage and manipulation.
- Add 10-15 ml of the preheated (50 °C) TE buffer to each tube containing the plugs. Ensure proper sealing of the tubes and shake them in the 50 °C shaker incubator for 10-15 minutes. The purpose of this step is to remove any residual contaminants and further purify the plugs.
- Carefully pour off the TE buffer from the tubes, ensuring that the plugs remain intact. Repeat the wash process with TE buffer for a total of four times, using fresh buffer each time. This ensures thorough washing and removal of any remaining impurities.
- After the final wash, transfer the plugs to microcentrifuge tubes containing TE buffer. Store 2-4 plugs per tube and keep them at 4 °C until they are needed for further analysis or processing. Storing the plugs in TE buffer helps maintain their integrity and prevents DNA degradation.
Proper washing of the agarose plugs is crucial to remove contaminants and prepare them for subsequent steps in the PFGE workflow, such as DNA digestion and electrophoresis.