Proteins, as complex biological molecules, play an essential role in the myriad of processes in living organisms. Consisting of amino acids, proteins can be envisioned as polypeptide structures formed through long chains of these amino acid residues. Therefore, it’s not surprising that they are one of the most abundant organic molecules present in living beings. Besides acting as the structural backbone for organisms, proteins serve as catalysts, enzymes, hormones, and vital regulators of cellular mechanisms.
Understanding the intricacies of protein structure is pivotal. Why? Because the structure of a protein, specifically its three-dimensional arrangement of atoms, largely dictates its function. Furthermore, proteins are unique. In any given living system, there are thousands of different proteins, each tailored for a specific function determined by its unique structure.
Diving deeper into the architectural complexity of proteins, one finds that proteins are comprised of several layers of structure. At its core, every protein possesses a primary structure, a linear sequence of amino acids. Then, this primary structure gives way to a secondary structure, where regions of amino acid chains stabilize via hydrogen bonds. These bonds, in turn, produce patterns known as alpha-helix and beta-pleated sheets. Progressing further, the tertiary structure emerges, influenced by the interactions of side chains from the polypeptide backbone. Lastly, although not applicable to all proteins, some exhibit a quaternary structure, which results from the interactions between two or more polypeptide chains.
An interesting aspect to consider is the polymer nature of proteins. Proteins are essentially polymers, more specifically termed as polypeptides. These are sequences derived from amino acids, which can be visualized as the fundamental building blocks or monomers. These amino acids, when they undergo condensation reactions, give rise to peptides, and by extension, proteins. It’s a delicate dance of molecular interactions – from hydrogen bonding and ionic interactions to Van der Waals forces and hydrophobic packing – that ensures proteins fold into their specific spatial conformations.
To decipher the mysteries of protein functions at such a molecular scale, their three-dimensional structure needs to be determined. This necessity has given birth to the field of structural biology. Techniques like X-ray crystallography, NMR spectroscopy, and cryo-electron microscopy (cryo-EM) are employed within this scientific domain to unveil the intricate structure of proteins.
It’s imperative to understand that proteins can range in size, with some having mere tens of amino acids and others comprising several thousand. By dimension, proteins fall under the category of nanoparticles, specifically measuring between 1–100 nm. In certain instances, protein complexes, vast in size, can emerge from subunits, much like how numerous actin molecules come together to form a microfilament.
Furthermore, proteins are dynamic. In executing their biological roles, they often undergo structural modifications. These alternative structures, or conformations, are crucial for a protein’s function, and the shifts between them are termed conformational changes. In conclusion, the realm of proteins is vast, intricate, and indispensable for life, warranting continued exploration and understanding.
Levels of Protein Structure
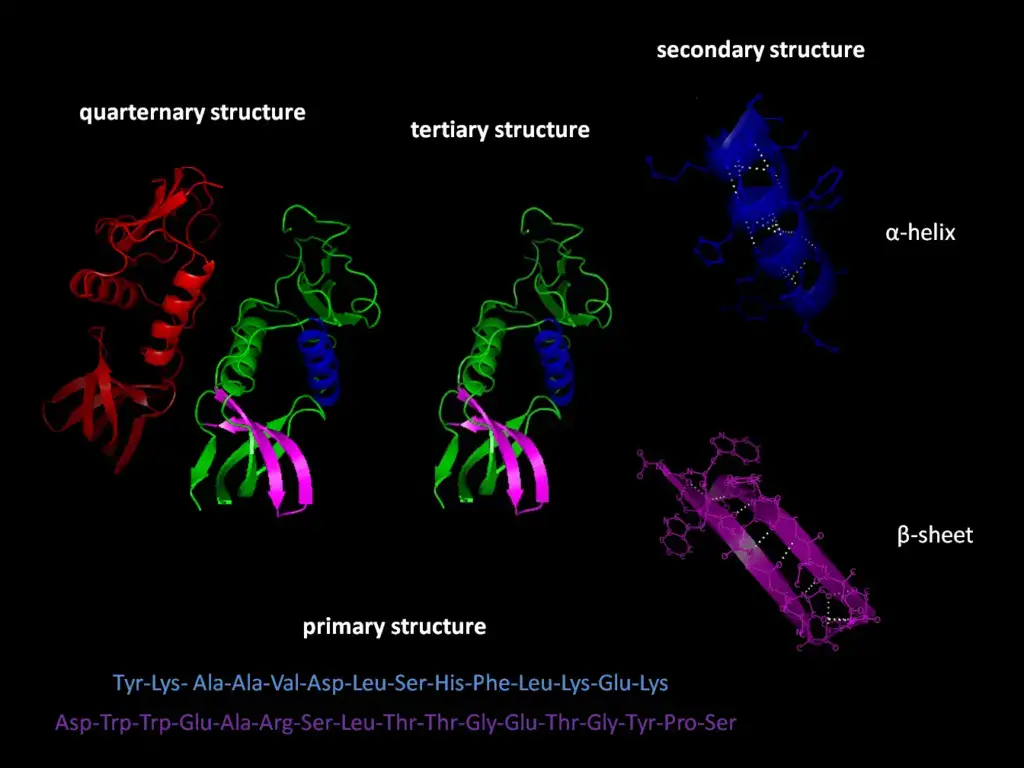
Proteins, being complex biological molecules, possess intricate structural designs that facilitate their diverse functions within living organisms. Understanding the structural levels of proteins is crucial to comprehend their functions and behavior. Therefore, one can identify four distinct levels of protein structure:
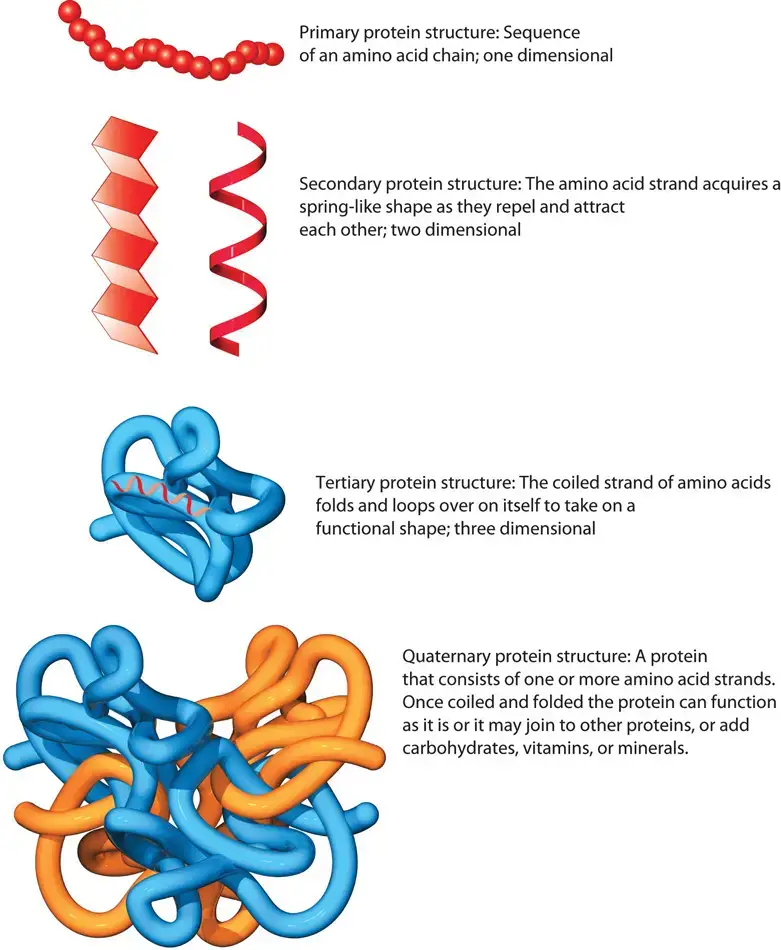
- Primary Structure: The foundation of protein structure lies in its primary structure, which represents the one-dimensional sequence of amino acids in a polypeptide chain. These amino acids are connected by peptide bonds. This sequence determines how the protein will fold in the subsequent structural levels.
- Secondary Structure: Moving beyond the linear sequence, the secondary structure arises when the polypeptide chain assumes specific configurations. This formation, which can be in the form of coils or helices and sheets, is governed by the chemical interactions between the amino acids. At this level, patterns like alpha-helices and beta-sheets emerge due to these interactions.
- Tertiary Structure: Delving deeper, the tertiary structure reveals the unique three-dimensional shape of the protein. This structure is an outcome of the interactions between the various amino acid side chains. Some side chains may attract each other, while others may repel, leading the protein to fold into a specific shape. Therefore, the sequence of amino acids in the primary structure ultimately influences the protein’s tertiary formation.
- Quaternary Structure: The final level, the quaternary structure, is observed in proteins that consist of multiple polypeptide subunits. These subunits come together to form a single, functional protein. An illustrative example of a protein with a quaternary structure is hemoglobin, which is composed of four polypeptide chains that bond together, serving as an oxygen carrier in the bloodstream.
Besides its biological functions, a protein’s structure significantly influences its nutritional quality. For instance, proteins with large fibrous structures pose challenges during digestion compared to smaller ones. Some proteins, like keratin, are even indigestible. As a result, not all amino acids from such proteins are absorbed, reducing their nutritional value.
However, proteins are sensitive to their environment. Alterations in their physical conditions can disrupt their three-dimensional structure, a phenomenon known as denaturation. Denaturation leads to both the loss of structural integrity and functional ability of proteins. Various factors, such as changes in pH, exposure to heavy metals, alcohol, or heat, can induce protein denaturation. This process is observable in everyday life; for instance, the transformation of liquid egg whites into a solid form when cooked or the fluffiness of whipped cream. Both scenarios highlight the denaturation causing physical changes in protein structure. Since the structure of a protein is closely linked to its function, denaturation inevitably impacts its functionality. In conclusion, the multifaceted structural levels of proteins dictate their functions, nutritional value, and reactions to environmental changes.
1. Primary Structure of Protein
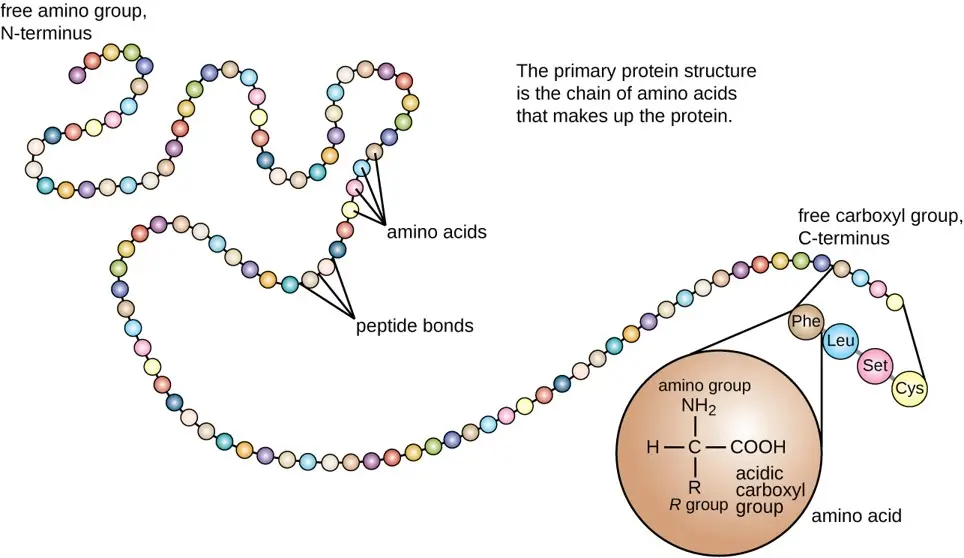
- The primary structure of a protein is the most fundamental aspect of its architecture, denoting the linear sequence of amino acids within a polypeptide chain. These amino acids are linked by covalent peptide bonds, and the sequence is directly encoded by the organism’s genetic material. Therefore, the primary structure can be perceived as a direct consequence of the nucleotide sequence present in the DNA, transcribed into mRNA, and subsequently translated by ribosomes into the polypeptide chain.
- Each amino acid in the sequence is unique, characterized by a central alpha carbon atom bonded to four different groups: a hydrogen atom, a carboxyl group, an amino group, and a variable side chain, denoted as the “R” group. It is this “R” group that imparts distinct chemical properties to each amino acid, influencing the protein’s overall folding and function. The sequence of amino acids in the primary structure is often referred to in terms of “residues,” considering that in the formation of a peptide bond, a water molecule is released, thus the product is a residue of the original amino acid.
- The two ends of the polypeptide chain are the carboxyl terminus (C-terminus) and the amino terminus (N-terminus), with the residue numbering beginning from the N-terminal end. The specificity of the primary structure is such that even a single change in the amino acid sequence due to a gene mutation can lead to a drastic alteration in the protein’s properties. This is exemplified in genetic disorders like sickle-cell anemia, where a single amino acid substitution can affect the protein’s function—in this case, hemoglobin.
- Furthermore, it is important to consider post-translational modifications as part of the primary structure, though these modifications occur after the initial synthesis of the polypeptide chain and are not encoded by the gene. Such modifications include phosphorylations and glycosylations, which can modify the behavior and function of the protein.
- The elucidation of the primary structure of a protein can be achieved through techniques such as Edman degradation or tandem mass spectrometry. Frederick Sanger’s pioneering work on insulin was a landmark in establishing that proteins have a definitive amino acid sequence, setting the stage for the understanding that the sequence dictates the structure and function of the protein.
- In conclusion, the primary structure of a protein serves as the blueprint for the protein’s final three-dimensional shape and function. It is a precise and unambiguous sequence of amino acid residues, determined by the genetic code, and held together by peptide bonds. The integrity of this sequence is vital, as any alteration can have profound implications on the protein’s role within a biological system.
Peptide Bond Formation and Primary Protein Structure
Proteins are vital macromolecules found within cellular systems. Their synthesis is orchestrated by ribosomes, complex enzymes comprising RNA and proteins. The order in which amino acids are arranged to form a protein is called its primary sequence, governed by the genetic data present in the messenger RNA (mRNA) molecule.
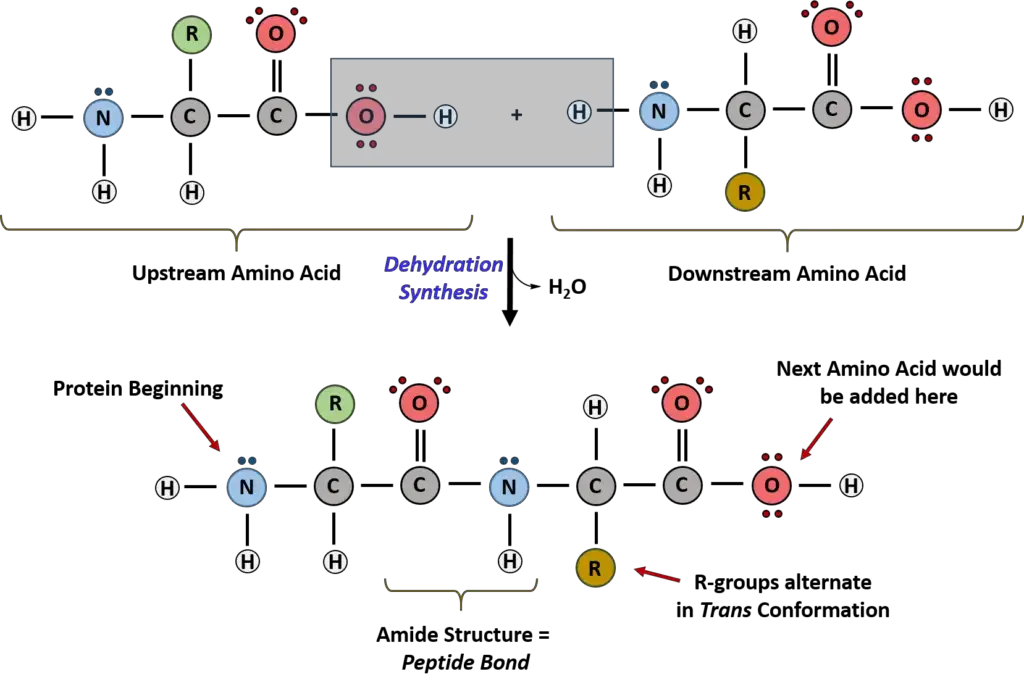
- Dehydration Synthesis in Protein Formation
- Dehydration synthesis is a crucial process that facilitates the connection of amino acids to form proteins. This process involves the removal of water and combines the carboxylic acid of one amino acid with the amine functional group of the subsequent amino acid, forming an amide linkage.
- In contrast, hydrolysis involves the addition of water to break the amide bond between amino acids. This reverse reaction is facilitated by enzymes known as proteases.
- Understanding the Peptide Bond
- The specific linkage formed between amino acids in protein structures is termed the peptide bond.
- Protein synthesis is always directional, commencing from the amine and culminating at the carboxylic acid tail. The ribosome ensures that new amino acids are added only to the carboxylic acid end, thus confirming an N- to C- synthesis.
- Amide Resonance and Its Implications
- Amide bonds exhibit a resonance structure, preventing the nitrogen’s lone pair of electrons from acting as a base. Instead, these electrons form a pi-bond with the carbonyl carbon, thereby establishing a double bond. Consequently, the C-N bond within the amide is spatially fixed due to this pi-bond character.
- The implications of this are profound. The fixed spatial position leads to a distinct physical arrangement of the R-groups in either cis or trans conformations. Typically, the trans conformation is more common due to reduced steric hindrance, except in the case of the amino acid proline, which favors the cis conformation because of its unique cyclic structure.
- Peptides Versus Proteins
- Proteins are essentially long chains of amino acids. Macromolecules that are smaller than 50 amino acids are referred to as peptides.
- The number of possible protein structures is astronomical due to the numerous combinations of amino acids. The sequential arrangement of these amino acids significantly influences the 3-dimensional structure and, therefore, the function of the protein.
- Characteristic Features of Amino Acids
- The inherent properties of amino acids govern how a protein folds to acquire its 3-dimensional shape. This shape is crucial for its functional activities.
- For proteins located within the cell’s aqueous environment, hydrophobic amino acids are typically situated internally, while hydrophilic amino acids interface with external water molecules. Proline, because of its cyclic structure, often indicates bends or changes in protein direction.
- Methionine is distinctive, serving as the starting amino acid for most proteins. Moreover, cysteines, with their thiol functional groups, can oxidize to form covalent disulfide bonds, lending added stability to the protein’s structure.
- The Role of Disulfide Bonds
- Disulfide bonds form between two cysteine residues within a peptide or protein sequence or even between distinct peptide or protein chains. For instance, the hormone insulin requires disulfide bridges between its peptide chains for proper functioning and regulating blood glucose levels.
2. Secondary Structure of Protein
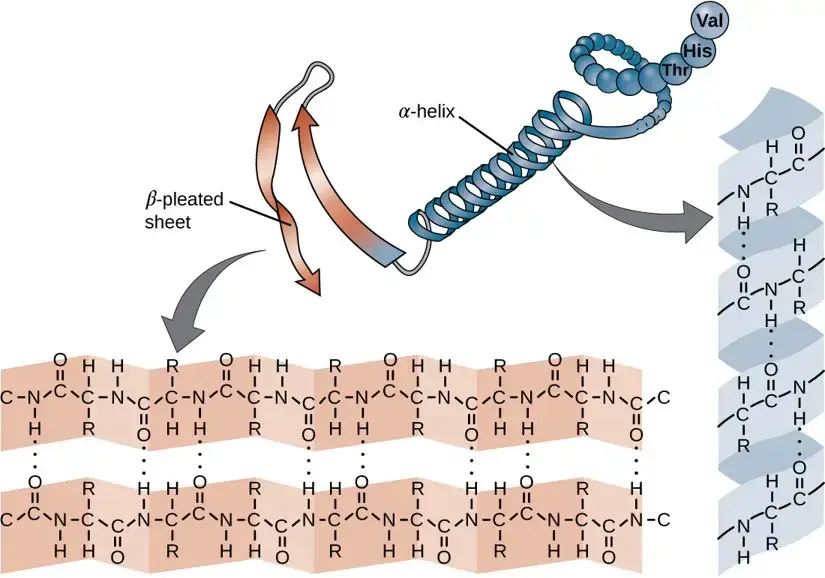
The secondary structure of a protein refers to the conformation adopted by the polypeptide chain due to twisting or folding. In this configuration, the amino acids lie close to each other in their sequence. There are primarily two recognized secondary structures: the α-helix and the β-sheet. Ramachandran, an Indian scientist, made a significant contribution in understanding the spatial arrangement of these polypeptide chains.
- α-Helix
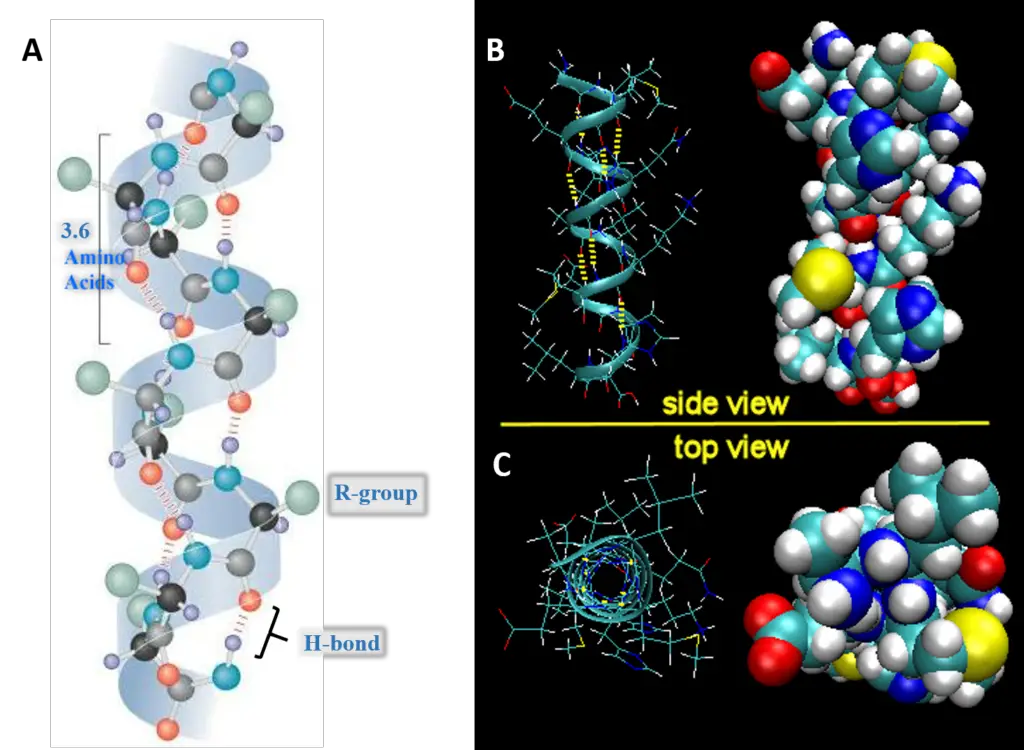
- Definition and Discovery:The α-helix is a commonly found spiral structure in proteins. Proposed by Pauling and Corey in 1951, its identification is considered a milestone in biochemistry research.
- Characteristics:
- It is a tightly packed coiled structure with amino acid side chains projecting outward from a central axis.
- Stability in the α-helix is due to extensive hydrogen bonding, formed between the H atom of a peptide N and the O atom of a peptide C. Though these hydrogen bonds are weak individually, their collective strength stabilizes the helix.
- Almost all the peptide bonds in a polypeptide chain, barring the first and last, partake in hydrogen bonding.
- Each turn of the α-helix comprises 3.6 amino acids and covers 0.54 nm. There is a 0.15 nm spacing between each amino acid.
- The α-helix adopts a stable conformation with the lowest energy.
- A right-handed α-helix is more stable than its left-handed counterpart.
- Certain amino acids, notably proline, disrupt the α-helix. Large quantities of acidic (e.g., Asp, Glu) or basic (e.g., Lys, Arg, His) amino acids can also interfere with this structure.
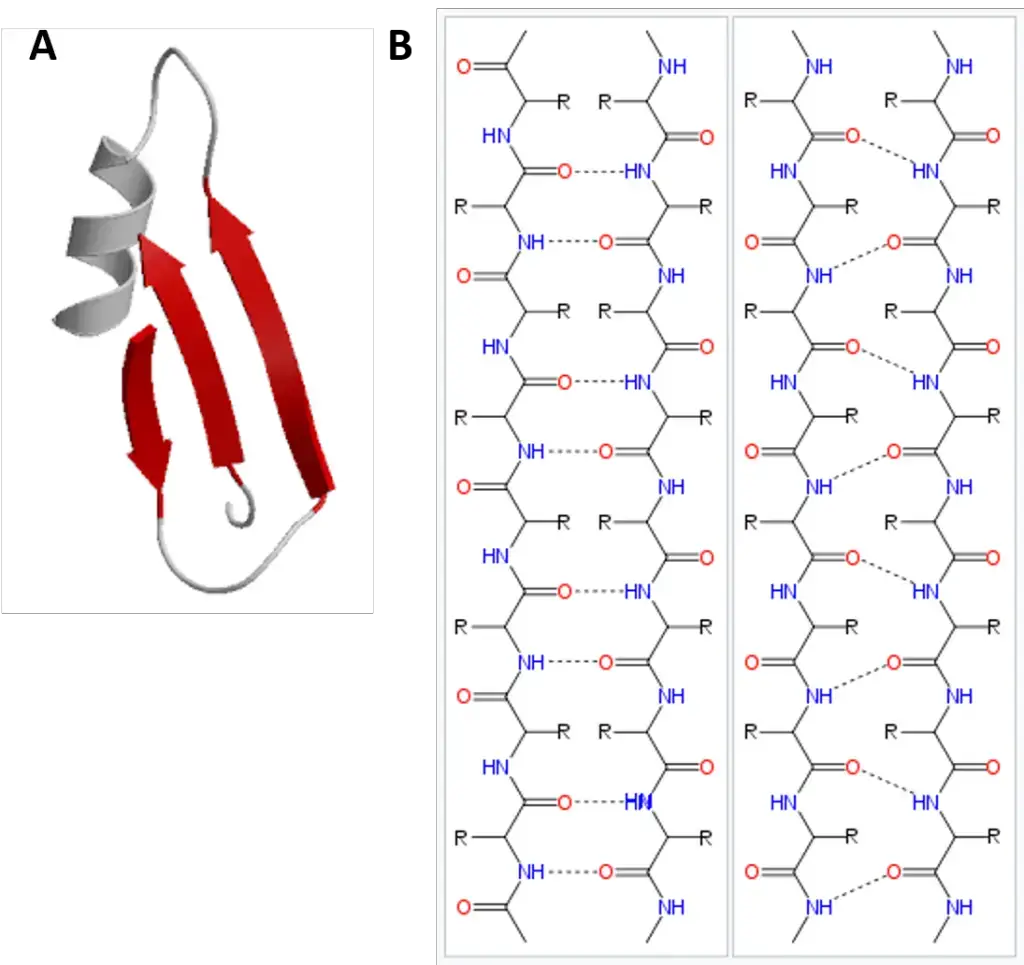
- β-Pleated Sheet
- Definition and Discovery:The β-pleated sheet is the second form of protein secondary structure, as proposed by Pauling and Corey.
- Characteristics:
- The β-pleated sheets consist of two or more fully extended peptide chain segments.
- Hydrogen bonds between neighboring peptide chain segments stabilize the structure.
- Types:Parallel and Anti-parallel β-Sheets: In β-sheets, polypeptide chains can be arranged either parallel (same direction) or anti-parallel (opposite direction). The formation of β-pleated sheets can occur either between separate polypeptide chains (interchain H-bonds) or within a single chain that folds back on itself (intrachain H-bonds).
- Occurrence:A variety of proteins feature β-pleated sheets. Notably, the α-helix and β-sheet structures often coexist in the same protein, with β-sheets frequently forming the core of globular proteins.
- Other Secondary Structures:Besides the aforementioned α-helix and β-sheet structures, proteins can also exhibit β-bends and non-repetitive, less organized secondary structures.
3. Tertiary Structure of Protein
The tertiary structure of a protein refers to its intricate three-dimensional conformation. This structure emerges from the folding of the secondary structural elements, such as α-helices and β-pleated sheets, into a compact globular form.
- Composition and Stability
- Hydrophobic and Hydrophilic Arrangement:In the tertiary structure, hydrophobic side chains are typically oriented towards the interior of the protein, while hydrophilic groups are exposed to the aqueous environment, enhancing the molecule’s stability.
- Bonding Interactions:The structure is stabilized through various interactions, including hydrogen bonds, disulfide bonds (S-S), ionic interactions (also known as electrostatic bonds), hydrophobic interactions, and van der Waals forces.
- Domains
- Definition and Functionality:A domain is a fundamental unit within the tertiary structure that contributes to the protein’s function. It is a discrete, locally folded region of the protein that typically contains 100–200 amino acids.
- Domain Structure and Importance:Domains are tightly packed, globular configurations that maintain structural integrity and facilitate the protein’s functional role. They are often connected by loop regions, which allow for flexibility and interaction with other molecular entities.
- Further Folding of Secondary Structures
- Formation of Tertiary Structure:The tertiary structure arises from the further folding and twisting of secondary structures, driven primarily by the side-chain interactions of the amino acids in the polypeptide backbone.
- Types of Molecular Shapes:The arrangement of these folds results in two principal molecular configurations: fibrous and globular shapes, which dictate the protein’s overall structure and function.
- Stabilizing Forces
- Detailed Interactions:The secondary and tertiary structures are maintained by several forces, including hydrogen bonds, disulfide linkages, van der Waals interactions, and electrostatic forces of attraction.
- Specificity of Interactions:While non-specific hydrophobic interactions play a role in the initial folding process, the precise tertiary structure is locked in place by more specific interactions such as salt bridges, the close packing of side chains, and disulfide bonds.
- Environment Influence on Tertiary Structure
- Disulfide Bond Occurrence:Disulfide bonds, which can play a critical role in stabilizing the tertiary structure, are less common in proteins within the cytosol due to its generally reducing environment.
4. Quaternary Structure of Protein
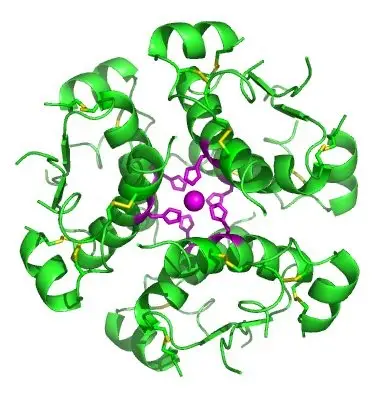
The quaternary structure of proteins is characterized by the assembly of multiple polypeptide chains or subunits. These subunits, which can be identical or different, are also known as monomers, protomers, or subunits. The proteins composed of two or more polypeptides are termed oligomers.
- Classification of Subunit Assemblies
- Dimeric and Tetrameric Structures:When two polypeptides associate, the structure is called a dimer, while an assembly of four subunits is termed a tetramer. The number of subunits dictates the multiplicity of the oligomeric protein.
- Bonding in Quaternary Structure
- Types of Bonds:The subunits in a quaternary structure are held together through non-covalent interactions, which include hydrogen bonds, hydrophobic interactions, and ionic bonds.
- Functional Significance of Oligomeric Proteins
- Metabolic and Cellular Roles:Oligomeric proteins are integral to regulating metabolism and other cellular functions, demonstrating the importance of quaternary structure in biological systems.
- Exemplification of Oligomeric Proteins
- Representative Examples:Hemoglobin, aspartate transcarbamoylase, and lactate dehydrogenase are prime examples of oligomeric proteins that exhibit quaternary structure.
- Spatial Arrangement of Subunits
- Composition of Quaternary Structure:The quaternary structure arises from the spatial arrangement of individual tertiary structures, which are the polypeptide chains that are organized in specific three-dimensional shapes.
- Characterization of Quaternary Structure
- Definition and Complexity:The quaternary structure is not merely a congregation of polypeptide chains but a complex spatial arrangement of these subunits. This structure is the highest level of organization observed in protein architecture.
- Variability and Complexity:Ranging from simple dimers to complex multisubunit configurations, the quaternary structure can involve an intricate interplay of multiple tertiary structures.
- Structural and Functional Integration
- Interactions Among Subunits:Each subunit possesses its unique primary, secondary, and tertiary structures that interact via hydrogen and van der Waals forces, contributing to the stability and functionality of the entire protein complex.
- Precision in Subunit Organization:The precise organization of subunits is paramount. Deviations in their arrangement can lead to significant alterations in protein function, highlighting the delicate interdependence of structure and function in quaternary protein complexes.
Protein Folding Process
- Initiation: Polypeptide Synthesis and Early Folding
- As soon as a polypeptide chain starts its synthesis by a ribosome, it begins the protein folding process. The initial sequence of amino acids determines the eventual shape the protein will take.
- Primary Structure’s Influence
- The primary structure of a protein is its linear sequence of amino acids. This sequence is encoded by the mRNA that the ribosome translates. The specific order of amino acids in this chain, as governed by Anfinsen’s dogma, dictates the protein’s ultimate folded structure and thus its function.
- Formation of Secondary Structures
- Driven by the hydrogen bonds between peptide bond components, the chain forms alpha helices and beta sheets. These structures serve as the protein’s backbone, providing initial shape and rigidity.
- Tertiary Structure Development
- As the protein continues to fold, further interactions occur:
- Hydrophobic interactions: Amino acids with hydrophobic (water-repellent) side chains tend to move away from the aqueous environment and cluster together in the protein’s core.
- Ionic bonds: Between positively and negatively charged side chains of different amino acids.
- Disulfide bridges: Covalent bonds that form between sulfur atoms of two cysteine amino acids.
- Van der Waals interactions: Weak attractions between close atoms.
- The culmination of these interactions results in the protein adopting its three-dimensional structure or tertiary structure.
- As the protein continues to fold, further interactions occur:
- Quaternary Structure Assembly (If Applicable)
- Some proteins consist of multiple polypeptide chains (subunits). The spatial arrangement of these subunits with respect to one another forms the quaternary structure of the protein.
- Role of External Factors
- External factors, including solvent interactions (usually water), pH, temperature, ionic concentration, and the presence of molecular chaperones, can influence the folding process. For instance, molecular chaperones assist proteins in achieving their native state by preventing incorrect folding or promoting correct folding pathways.
- Co-translational vs. Post-translational Folding
- Co-translational folding refers to folding that happens concurrently with translation, where the N-terminus of the protein begins to fold even as the remaining part is still being synthesized by the ribosome.
- Post-translational folding occurs after the polypeptide chain has been completely synthesized.
- Functional Testing and Refinement
- Once the protein achieves its native state, it undergoes “functional testing” where it interacts with other cellular components. If functional, the protein remains in its current conformation; otherwise, it may undergo further refinements or be targeted for degradation.
- Misfolding and Its Consequences
- Sometimes proteins do not fold correctly. Misfolded proteins might become non-functional or acquire toxic functionalities. Accumulation of certain misfolded proteins is associated with diseases, such as Alzheimer’s disease, where amyloid fibrils form.
- Energy Landscape and Thermodynamics
- The protein folding process is thermodynamically favorable, resulting in a decrease in the system’s Gibbs free energy. This energetically favorable process ensures that proteins usually achieve their functional, native structure from an ensemble of unfolded states.
Determination of primary structure of Protein
The primary structure of a protein refers to the unique linear sequence of amino acids that constitutes the protein. It offers essential insights into the protein’s function, interactions, and evolution. Detailed below is a systematic explanation of the stages involved in determining the primary structure of a protein.
The primary structure involves identifying the amino acids regarding their quality, quantity, and sequence within a protein structure. A purified protein or polypeptide sample is pivotal for this determination.
- Determination of Amino Acid Composition
- Protein Hydrolysis: A protein or polypeptide undergoes complete hydrolysis to release its constituent amino acids. This hydrolysis can be effected using acid, alkali, or enzymatic methods. Notably, enzymatic treatments, such as with pronase (a blend of non-specific proteolytic enzymes), often yield smaller peptides as opposed to individual amino acids.
- Separation and Estimation: Following hydrolysis, the liberated amino acids are differentiated and quantified using chromatographic techniques. Therefore, an understanding of chromatography becomes vital in grasping the nuances of protein primary structure determination.
- Degradation of Protein into Smaller Fragments
- Liberation of Polypeptides: Agents like urea or guanidine hydrochloride are employed to break non-covalent bonds, dissociating the protein into its constituent polypeptide chains. Moreover, to cleave the disulfide linkages binding the polypeptide units, treatment with performic acid becomes essential.
- Identification of Polypeptide Numbers: Dansyl chloride interacts specifically with the N-terminal amino acids of proteins, resulting in dansyl polypeptides. On subsequent hydrolysis, these yield N-terminal dansyl amino acids. Then, the count of these dansyl amino acids corresponds to the number of polypeptide chains present in the protein.
- Breakdown into Fragments: The larger polypeptides are further broken down into more manageable fragments through enzymatic or chemical methods.
- Enzymatic Cleavage: Enzymes such as trypsin, chymotrypsin, pepsin, and elastase are known for their specificity in cleaving peptide bonds. Trypsin, for instance, predominantly hydrolyzes peptide bonds associated with lysine or arginine, targeting the carbonyl side.
- Chemical Cleavage: Compounds like Cyanogen bromide (CNBr) are instrumental in fragmenting polypeptides. Specifically, CNBr targets peptide bonds where the carbonyl side is associated with the amino acid methionine.
- Determining the Amino Acid SequenceOnce the protein is fragmented into smaller peptides, these segments become suitable for sequence determination. This process is executed in a sequential manner.
- Sanger’s Reagent: Fred Sanger employed 1-fluoro 2,4-dinitrobenzene (FDNB) for ascertaining the insulin structure. This reagent attaches exclusively to the N-terminal amino acid, creating a dinitrophenyl (DNP) peptide derivative. Chromatography can then be used to identify the resulting DNP-amino acid. Nevertheless, the utility of Sanger’s reagent is restricted, mainly because it hydrolyzes the peptide chain fully into amino acids.
- Edman’s Reagent: Phenyl isothiocyanate, known as Edman’s reagent, interacts with the N-terminal amino acid to produce a phenyl thiocarbamyl derivative. A subsequent mild acid treatment releases a cyclic compound, phenyl thiohydantoin (PTH)–amino acid, which can then be identified via chromatography. Edman’s reagent possesses a distinct advantage since it sequentially degrades a peptide, releasing only the N-terminal amino acids without entirely hydrolyzing the peptide.
- Sequenator: An advanced instrument, the sequenator automates the process of determining the amino acid sequence in polypeptides (typically up to around 100 residues). Based on the principle of Edman’s degradation, this machine identifies amino acids sequentially from the N-terminal end. The liberated PTH-amino acid is recognized using high-performance liquid chromatography (HPLC), with each amino acid taking approximately two hours for determination.
Different Methods for Determination of protein structure
Proteins, with their intricate and diverse structures, play pivotal roles in various biological processes. Therefore, it becomes imperative to study and understand their spatial arrangements. A variety of techniques have been developed to elucidate protein structures. Here, we will explore these methods in a detailed and sequential manner.
- X-ray Crystallography
- Overview: X-ray crystallography stands as a prominent method in determining protein structures. It offers insights into the atomic arrangement within proteins and other crystallizable molecules.
- Process: The method encompasses the crystallization of the protein, following which it is exposed to X-rays. The diffraction pattern resulting from this exposure is then scrutinized to formulate an electron density map. Subsequently, this map assists in pinpointing the location of each atom within the protein.
- Applications and Limitations: While X-ray crystallography furnishes comprehensive atomic details, it is ideally suited for rigid proteins that can form well-ordered crystals. Flexible proteins pose challenges due to the necessity for precise molecular alignment.
- NMR Spectroscopy
- Overview: Nuclear Magnetic Resonance (NMR) spectroscopy is an invaluable tool for discerning protein structures in solution, eliminating the need for crystallization.
- Process: The procedure involves subjecting purified proteins to a magnetic field and subsequently probing them with radio waves. The resonances that emerge from this interaction divulge information about atomic conformation, culminating in a model of the protein structure.
- Applications and Limitations: NMR proves exceptionally beneficial for examining flexible proteins in solution. However, studying large proteins might introduce complexities due to overlapping peaks in the NMR spectra.
- Electron Microscopy (3DEM)
- Overview: Electron microscopy, especially the 3D Electron Microscopy (3DEM) technique, facilitates the determination of 3D structures of voluminous molecules.
- Process: This method operates by imaging biomolecules using electron beams and lenses. A prevalent approach, Cryo-EM, involves capturing individual particles in ice, resulting in 2D projection images. These projections undergo computational processing to yield 3D density maps, into which atomic models are fitted for interpretation.
- Advancements: The recent surge in advancements across sample preparation, detectors, and data processing has augmented the efficacy of 3DEM.
- Computational Tools and Homology Modeling
- Overview: Experimental data, such as that obtained from X-ray diffraction patterns, NMR atomic details, or electron microscopy, at times, might not be adequate for devising an exhaustive atomic model. In such instances, computational tools come to the fore.
- Homology Modeling: Among these tools, homology modeling stands out. It predicates the three-dimensional structure of a protein based on a known protein structure, termed as a template. By juxtaposing the amino acid sequences of the target protein with the template, a model of the target protein’s structure is constructed.
- Partial Structural Study Methods: Understanding protein structures isn’t always about complete 3D visualizations. Sometimes, partial structural information can be invaluable.
- Techniques: These include ultracentrifugation, mass spectrometry, and fluorescence spectrometry.
- Function: These methods often complement other techniques, providing auxiliary data that enriches the understanding of protein structure.
References
- Alberts B, Johnson A, Lewis J, et al. Molecular Biology of the Cell. 4th edition. New York: Garland Science; 2002. Analyzing Protein Structure and Function. Available from: https://www.ncbi.nlm.nih.gov/books/NBK26820/
- Sanvictores T, Farci F. Biochemistry, Primary Protein Structure. [Updated 2022 Oct 31]. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2023 Jan-. Available from: https://www.ncbi.nlm.nih.gov/books/NBK564343/
- Brunori, M. (2008). [Methods in Enzymology] Globins and Other Nitric Oxide-Reactive Proteins, Part B Volume 437 || Structural Dynamics of Myoglobin. , (), 397–416. doi:10.1016/S0076-6879(07)37020-1
- Protein Structure Determination – https://www.news-medical.net/life-sciences/Protein-Structure-Determination.aspx
- METHODS TO DETERMINE PROTEIN STRUCTURE – https://www.slideshare.net/SabahatAli9/methods-to-determine-protein-structure
- Littlechild, Jennifer Ann (2013). Introduction to Biological and Small Molecule Drug Research and Development || Protein structure and function. , (), 57–79. doi:10.1016/B978-0-12-397176-0.00002-9
- https://www.sigmaaldrich.com/IN/en/applications/protein-biology/protein-structural-analysis
- https://openoregon.pressbooks.pub/nutritionscience/chapter/6a-protein-structure/
- https://wou.edu/chemistry/courses/online-chemistry-textbooks/ch450-and-ch451-biochemistry-defining-life-at-the-molecular-level/chapter-2-protein-structure/#CH450-2.2
- https://pdb101.rcsb.org/learn/guide-to-understanding-pdb-data/methods-for-determining-structure
- https://www.rapidnovor.com/protein-structure-and-how-to-study-it/
- https://www.khanacademy.org/science/biology/macromolecules/proteins-and-amino-acids/a/orders-of-protein-structure
- https://biologydictionary.net/protein-structure/
- https://teachmephysiology.com/biochemistry/protein-synthesis/protein-structure/
- https://www.thoughtco.com/protein-structure-373563
- https://lubrizolcdmo.com/technical-briefs/protein-structure/