Acridine Orange staining is a fluorescent staining method used to visualize nucleic acids and acidic vesicles of the cell. It is a membrane-permeant dye, and it enters inside living cells where it binds with DNA and RNA. The staining is based on the metachromatic property of the dye because it emits different colours depending on the structure it is attached to.
When Acridine Orange binds with double-stranded DNA, it is present in monomeric form and it fluoresces green in the microscope. When it interacts with single-stranded RNA, it forms aggregates and the fluorescence becomes orange to red. It is the process in which the dye also accumulates inside acidic vesicular organelles (AVOs) such as lysosomes, because AO is a weak base and it becomes protonated in acidic pH and gets trapped, giving a bright red fluorescence.
This staining is the process used to study nucleic acid distribution, cell viability, apoptosis, autophagy, and growth phases of the cell cycle. The different colour emissions make it easy to differentiate nuclear DNA from cytoplasmic RNA and the acidic vesicles during microscopic observation.
Aim
Acridine Orange (AO) stain is a differential staining method of nucleic acids such as RNA and DNA.
Principle of Acridine Orange Staining
The principle of Acridine Orange staining is based on the metachromatic behaviour of the dye and its interaction with different nucleic acids and acidic compartments inside the cell. It is a weakly basic dye, and it can easily pass through the cell membrane to enter the cytoplasm and nucleus. When the dye binds with nucleic acids, the optical property of the dye is changed, and it begins to fluoresce under ultraviolet light. It is the process in which double-stranded DNA binds the dye by intercalation, keeping the molecules in monomeric form, so the fluorescence becomes green. This is referred to as the green emission of DNA. In contrast, when the dye interacts with single-stranded nucleic acids like RNA, the local concentration becomes high and the dye molecules form aggregates, resulting in orange to red fluorescence. The reaction is dependent on the structure and pH of the cellular components, and these are used to differentiate DNA and RNA during observation.
It is also the process where the dye accumulates inside acidic vesicular organelles because AO is a weak base, and it becomes protonated in low pH. In this condition the dye cannot diffuse back, and it gets trapped in lysosomes and similar organelles, which produces a bright red fluorescence due to heavy aggregation. This principle makes it useful to study nucleic acid distribution and the presence of acidic compartments in living cells.
Requirements
General Requirements
- It is important to use purified Acridine Orange dye for proper staining.
- The dye and stained sample is protected from light because AO is highly photosensitive.
- The concentration of the dye is kept same in every experiment as the colour shift is concentration-dependent.
- AO can be diluted in distilled water (diH₂O), PBS, or in media.
Requirements for Live Cell / Lysosomal Staining
- AO is added in the media at 0.5–5.0 μM for lysosomal structures.
- The cells are incubated for 15–30 minutes at 37°C.
- Gentle mixing is required so the cells are not damaged.
- Over-digestion with trypsin-EDTA is avoided because the dye can leak out of cells.
- For red fluorescence, excitation filter around 540–560 nm and emission >610 nm is used.
- For yellowish green fluorescence, excitation at 488 nm and emission around 540–550 nm is used.
- Washing of cells may be done if the slide appears too bright.
Requirements for Cell Cycle Analysis
- The staining is done under strictly controlled buffer chemistry and fixation.
- The medium is kept acidic with a final pH nearly 3.5 using citric acid (pH 3.0) and sodium phosphate (pH 3.8).
- Buffer #1 contains Triton X-100 (0.1%), sucrose (0.2M), and disodium EDTA (10⁻⁴ M).
- The AO working solution is freshly prepared by diluting the stock (like 2 mg/ml) to 20 μg/ml in Buffer #2.
- The steps are adding Buffer #1, short incubation, adding AO stain, and running immediately in flow cytometer.
Requirements for Microorganism / Smear Staining
- A thin smear is made on a clean slide and air-dried or heat-dried.
- The smear is fixed using methanol.
- The slide is flooded with AO stain (like 0.01% AO-F) for 2–3 minutes.
- The slide is rinsed properly with water or PBS.
- The staining is done under acidic pH so bacteria fluoresce orange and tissues appear yellow or green.
- Visualization is done under a fluorescent microscope.
Procedure of Acridine Orange Staining
I. General Procedure for Microorganism/Smear Staining (Microscopy)
This method is used for rapid detection of bacteria or fungi. It is carried out in acidic pH so that differential staining can be obtained.
Steps
- Prepare the smear– It is done by spreading a thin layer of the specimen on a clean grease-free slide using a sterile stick.
- Dry the smear– The slide is allowed to air dry naturally. It can also be placed on a heating block for quick drying.
- Fixation– The smear is fixed by flooding it with methanol. Excess alcohol is drained and the slide is left to dry.
- Staining – The slide is flooded with Acridine Orange stain (0.01% AO-F) for 2–3 minutes.
- Rinsing– Excess stain is drained and the slide is washed with tap water or PBS.
- Drying– The slide is air dried. It can also be blotted gently using filter paper.
- Visualization– It is observed under a fluorescent microscope. Under acidic pH bacteria appear orange while tissue materials appear yellow or green.
II. Acridine Orange Staining for Live Cells / Acidic Vesicular Organelles (AVO)
It is the process where AO accumulates in acidic vesicles like lysosomes due to its weak basic nature. These organelles show bright red fluorescence.
Steps
- Preparation of AO solution– AO may be used directly or diluted with PBS, media, or distilled water. Stock solution is generally supplied at 1 mM concentration.
- Dye dilution and addition– AO is added to cell media at a final concentration of 0.5–5.0 μM.
For 1 μM solution, stock is diluted 1:100 and then added to the cell suspension in 1:10 ratio. - Incubation– Cells are incubated for 15–30 minutes at 37°C. Some protocols use 5 μM AO in PBS for 60 minutes at 37°C.
- Protection from light– AO is photosensitive so the staining process is done protected from bright light.
- Washing (optional)– If staining appears very bright, the cells can be washed before viewing.
- Analysis– It is analyzed under fluorescence microscopy or flow cytometry.
- Red lysosomes are seen with 550 nm excitation and >610 nm emission.
- Yellow-green lysosomes appear with 488 nm excitation and 540–550 nm emission.
III. Acridine Orange Staining for Flow Cytometry Cell Cycle Analysis
It is used for quantifying DNA and RNA. This is the process requiring acidic buffers and precise timing.
Steps
- Cell preparation– About 10⁵–10⁶ cells are taken in 100 μL medium.
- Permeabilization and stabilization– 0.5 mL of Stock Buffer #1 is added. Buffer contains 0.1% Triton X-100, 0.2 M sucrose, 10⁻⁴ M EDTA in citrate phosphate buffer of pH ~3.5. Incubation is done for 1 minute.
- Preparation of staining solution– Fresh AO staining solution is prepared by mixing 0.1 mL AO stock (2 mg/mL) with 9.9 mL Stock Buffer #2 to make 20 μg/mL AO.
- Staining– 0.5 mL of freshly prepared AO stain is added to the sample.
- Analysis– It is run immediately on flow cytometer.
- Green fluorescence (DNA) is collected around 525 nm.
- Red/orange fluorescence (RNA) is collected around 650 nm.
IV. Dual Staining with Acridine Orange / Ethidium Bromide (AO/EB)
This staining is used for observing apoptosis and cell viability. It is the process where AO enters all cells but EB only enters non-viable or membrane-damaged cells.
Steps
- Cell preparation– About 2×10⁴ cells/mL are cultured under required conditions.
- Collection of cells– Cells are harvested, usually by trypsinization, and 25 μL suspension is placed on a clean slide.
- Dual staining– 1 μL AO/EB mixture (100 μg/mL AO + 100 μg/mL EB) is added to the suspension.
- Covering– A coverslip is placed gently over the drop.
- Microscopy– Cells are examined within 20 minutes using fluorescent microscope.
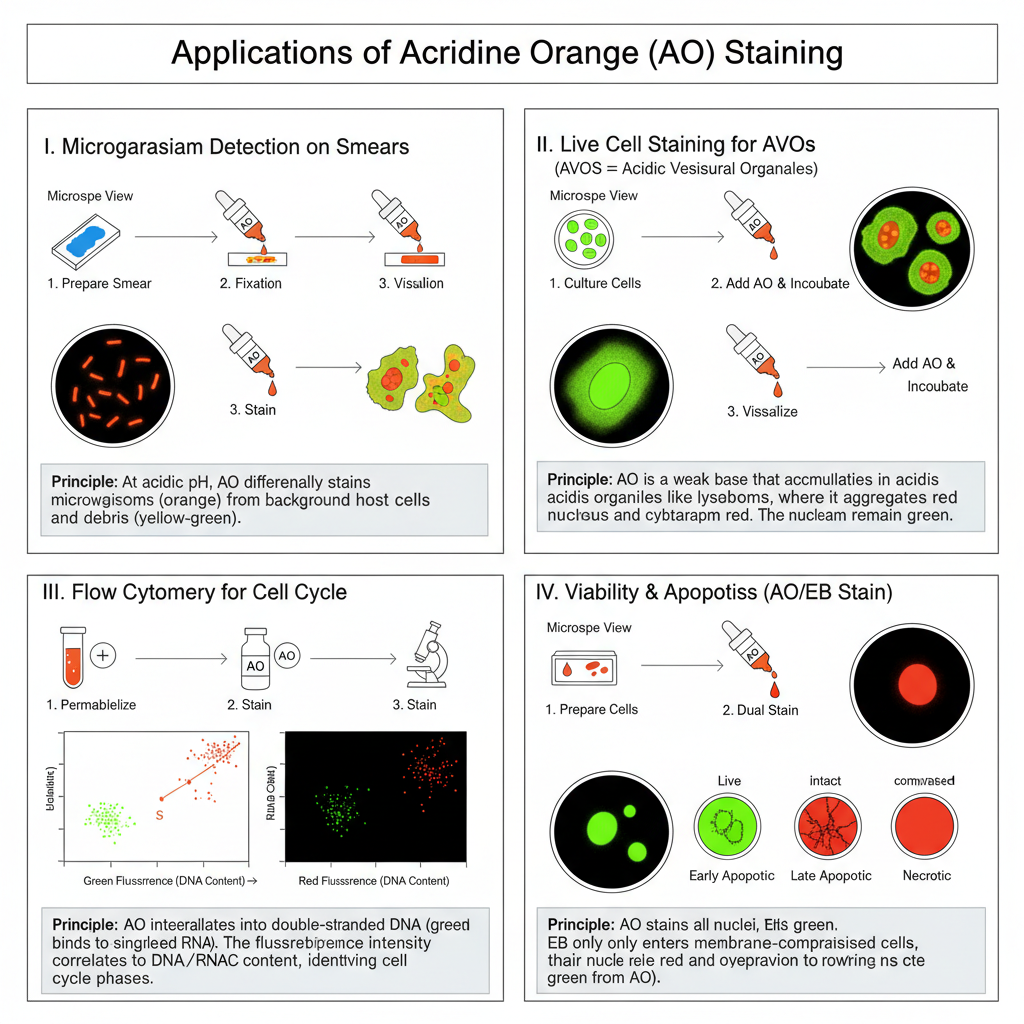
Observation of cell status
| Cell Type | Staining Result |
|---|---|
| Normal cells | Green uniform nucleus |
| Early apoptotic cells | Yellow-green AO staining, crescent or granular pattern, membrane intact |
| Late apoptotic cells | Orange EB staining, concentrated and asymmetrical, membrane compromised |
| Necrotic cells | Diffuse orange-red fluorescence, cell swelling or disintegration |
Result interpretation
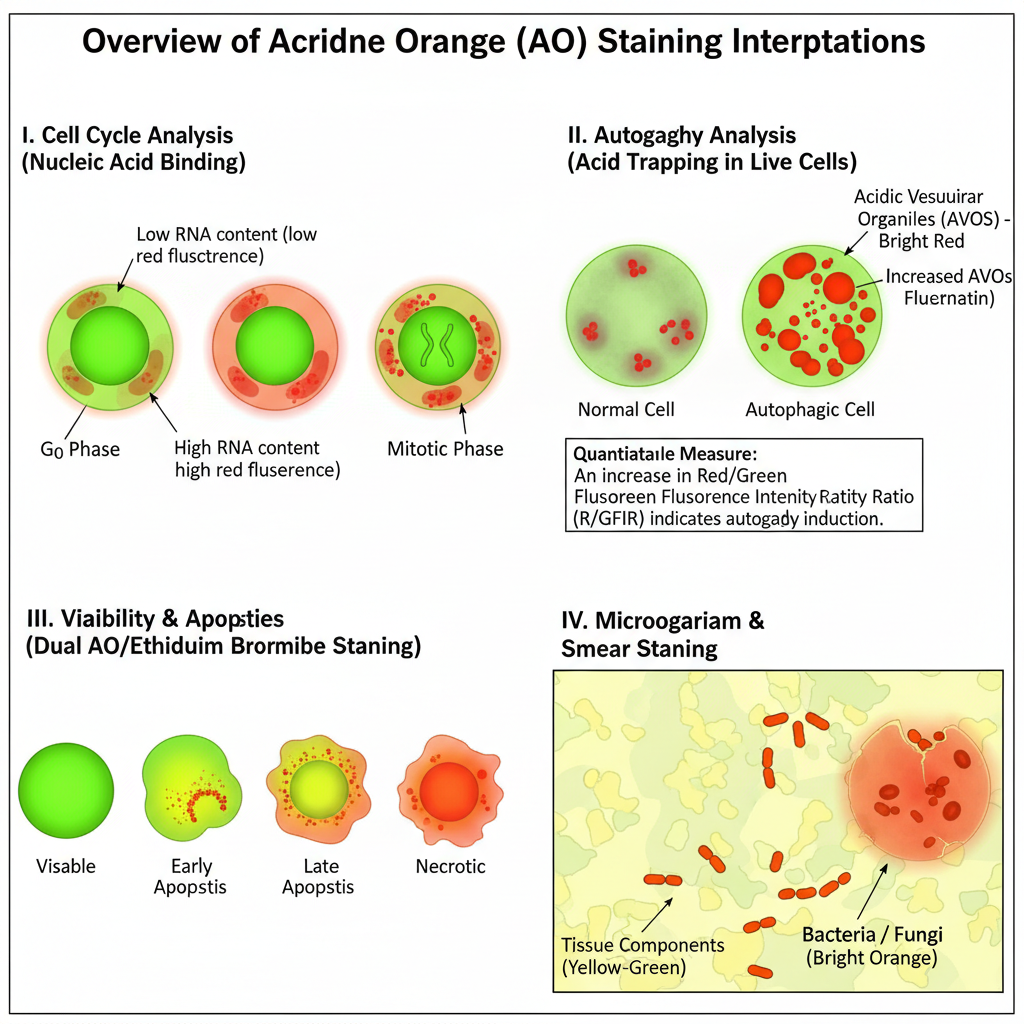
I. Interpretation Based on Nucleic Acid Binding (Cell Cycle / Quantification)
In this method, AO binds differently with double-stranded DNA (dsDNA) and single-stranded nucleic acids (ssRNA or ssDNA). It is the process widely used in cell cycle studies.
Some of the main interpretation points are–
- dsDNA shows green fluorescence at around 525 nm.
It is due to intercalation of AO in its monomeric form and this signal is proportional to DNA amount. - ssRNA or ssDNA shows red or orange fluorescence at around 630–650 nm.
It is due to electrostatic binding and formation of aggregates in high concentration.
In Cell Cycle Analysis
- G0 cells shows 2N DNA but have low RNA content; thus they give low red fluorescence.
- G1 cells also shows 2N DNA but have higher RNA content giving high red signal.
This helps in identification of G0 and G1 population. - Mitotic cells can be identified because the metaphase chromatin shows a different staining pattern compared to interphase cells.
II. Interpretation Based on Acid Trapping (Live Cell / Autophagy Studies)
In live cell staining, AO accumulates in acidic vesicular organelles (AVOs) due to its lysosomotropic nature. It is the process used to understand lysosomal function and autophagy.
Interpretation
- Lysosomes / Autolysosomes (AVOs):
They show bright red fluorescence. It occurs due to acid trapping and formation of aggregates. This red signal is proportional to the acidic vesicle volume. - Cytoplasm and Nucleus:
They show green fluorescence because AO remains in monomeric form in neutral pH. - Loss of red fluorescence:
It indicates inhibition or failure of lysosomal activity, such as neutralization of acidic pH by inhibitors (e.g. Bafilomycin A1).
R/GFIR (Red-to-Green Fluorescence Intensity Ratio)
This ratio is used for quantitative interpretation.
- Increase in R/GFIR shows increase in acidic vesicles and indicates autophagy induction.
- Decrease in R/GFIR shows reduction in autophagy or blockage of acidification.
- It is preferred because it corrects variations in dye uptake and cell size.
III. Interpretation in Viability and Apoptosis Assays (Dual AO / Ethidium Bromide Staining)
In this method AO enters all cells but EB enters only membrane-damaged cells. It is used to interpret viability and apoptotic stages.
Some of the important interpretation points are–
- Normal (Viable) Cells:
They show uniform green nucleus. EB is excluded because the membrane is intact. - Early Apoptotic Cells:
They show yellow-green nucleus with crescent or granular chromatin.
Membrane is mostly intact so EB is excluded. - Late Apoptotic Cells:
They show orange or red concentrated nucleus.
Membrane integrity is lost allowing EB entry and EB displaces AO. - Necrotic Cells:
They show diffuse orange-red fluorescence, increased cell volume and disintegration.
EB enters early without chromatin condensation which makes it different from apoptosis.
IV. Interpretation in Microorganism / Smear Staining
This staining is done at acidic pH. It is used for detecting bacteria and fungi in smears.
Some of the important observation are–
- Bacteria / Fungi: appear bright orange.
- Tissue components: appear yellow or green.
In special diagnostic applications like post-mortem study of myocardial infarction, normal myocardium gives golden brown fluorescence, while ischemic or anoxic cells show yellow or yellow-green fluorescence.
Uses of Acridine Orange Staining
- It is used in cell cycle and proliferation studies. AO measures DNA and RNA together and helps in identifying G0 and G1 cells and chromatin changes.
- It is used to monitor acidic vesicular organelles (AVOs). Lysosomes and autolysosomes show red fluorescence and it helps in studying autophagy induction.
- It is used to assess autophagic flux. The red-to-green fluorescence ratio (R/GFIR) is taken to understand changes in AVO volume.
- It is used for lysosomal visualization. AO staining shows lysosomes clearly under fluorescence microscopy and flow cytometry.
- It is used in apoptosis and necrosis detection. In dual staining (AO/EB or AO/PI), viable, apoptotic and necrotic cells can be differentiated by their color patterns.
- It is used in drug sensitivity studies. AO/EB staining helps in examining drug-induced apoptosis in tumor cells.
- It is used for detection of microorganisms. Bacteria show bright orange fluorescence at acidic pH giving quick detection in smears.
- It is used to observe fungal and microbial morphology. Distinctive structures of microorganisms can be visualized under a fluorescent microscope.
- It is used to detect nucleic acids like dsRNA. AO shows green fluorescence with double-stranded RNA and helps in gel visualization.
- It is used in emerging cancer imaging techniques. AO accumulates in tumor cells and is used for intraoperative fluorescence guidance.
- It is used in photodynamic and radiodynamic therapy studies. AO acts as a photosensitizer and helps in experimental cancer therapy when exposed to light or radiation.
- It is used in circulating tumor cell (CTC) screening. AO fluorescence helps to distinguish tumor cells from normal leukocytes in liquid biopsy.
- It is used in rapid tissue examination. AO staining with confocal microscopy provides quick tissue evaluation comparable to standard H&E staining.
Limitations of Acridine Orange Staining
- It is not highly specific for autophagy. It can accumulate in many acidic compartments and not only in autolysosomes, so AO alone cannot be used as a reliable autophagy marker.
- It binds with different nucleic acids. AO binds with both DNA and RNA, so the fluorescence signal depends on changes in both nucleic acids which may affect quantitative studies.
- It is highly photosensitive. The dye gets degraded rapidly by bright light and stained samples must be protected during handling.
- The staining is concentration dependent. The metachromatic color shift varies with dye concentration so maintaining equal concentration in all experiments is necessary.
- It requires specific buffer conditions. Live cell assays need preservation of pH gradient, and cell cycle assays require acidic buffers around pH 3.5 for proper staining.
- There is interference due to change in cell size. Cells with reduced size may take less dye giving false negative results while enlarged cells may give false positive red fluorescence.
- RNA excitation is inefficient under common laser settings. Standard flow cytometers use 488 nm excitation which is not optimal for RNA aggregate staining.
- EDTA exposure affects live cell staining. Excessive EDTA during cell preparation can increase permeability and cause loss of dye giving false results.
- It has safety-related concerns. Acridine dyes have shown mutagenic effects in some bacteria and early studies reported tumor formation in experimental animals.
- Limited clinical acceptance. There is insufficient long-term safety data so its clinical use in imaging and therapy remains limited.
Advantages of Acridine Orange Staining
- It shows metachromatic property. AO gives different fluorescence colors with DNA, RNA and acidic vesicles which makes it useful in many assays.
- It penetrates cells easily. Being a small membrane-permeant molecule, it enters live and fixed cells and stains nucleus and cytoplasmic organelles.
- It distinguishes dsDNA and RNA/ssDNA. AO stains double-stranded DNA green and stains RNA or single-stranded DNA orange or red which helps in structural identification.
- It is useful in cell cycle analysis. AO measures DNA and RNA together and helps in identifying G0, G1, S, G2 and M phases accurately.
- It can differentiate G0 and G1 cells. G1 cells show higher red signal due to more RNA, which standard DNA dyes cannot detect.
- It helps in identifying mitotic cells. Metaphase chromatin stains differently from interphase chromatin so mitotic index can be determined.
- It is useful in monitoring autophagy. Acidic vesicular organelles show bright red fluorescence and help in studying lysosomal activity.
- It supports ratiometric analysis (R/GFIR). The red-to-green ratio provides a quantitative measure of AVO volume correcting for dye uptake differences.
- It is useful in apoptosis and viability studies. In dual staining, AO/EB gives clear identification of viable, apoptotic and necrotic cells with distinct color patterns.
- It is simple and economical for tumor chemosensitivity studies. AO/EB staining provides morphological details similar to advanced flow cytometry methods.
- It is rapid for microorganism detection. Bacterial cells appear orange at acidic pH and can be identified quickly in clinical samples.
- It gives differential staining in smears. Human cells stain green and prokaryotes stain orange which helps in quick screening at lower magnification.
- It is useful for dsRNA detection in gels. AO shows green fluorescence with double-stranded RNA and helps in convenient gel visualization.
- It helps in circulating tumor cell (CTC) screening. AO staining shows high reproducibility and is less-invasive and lower cost than many other methods.
- It supports surgical fluorescence guidance. AO accumulates in tumor tissue due to high RNA and acidic conditions helping in identifying tumor regions during surgery.
- It is applicable in photodynamic and radiodynamic therapies. AO can act as a photosensitizer and supports innovative anticancer treatment studies.
- AAT Bioquest. (2024). What is the general procedure of acridine orange staining?
- Antibodies Incorporated. (n.d.). Acridine orange fluorescent staining solution | 6130.
- [No Author Provided]. (n.d.). Acridine orange staining: A comprehensive review of photophysical mechanisms, quantitative cellular analysis, and advanced applications in cell biology [Unpublished manuscript excerpt].
- Biotium. (n.d.). Acridine orange, 10 mg/mL in water (high purity) (AO).
- Byvaltsev, V. A., Bardonova, L. A., Onaka, N. R., Polkin, R. A., Ochkal, S. V., Shepelev, V. V., Aliyev, M. A., & Potapov, A. A. (2019). Acridine orange: A review of novel applications for surgical cancer imaging and therapy. Frontiers in Oncology, 9(925).
- Carver College of Medicine, Flow Cytometry. (n.d.). Acridine orange for cell cycle analysis | Flow cytometry. University of Iowa.
- Darzynkiewicz, Z., Traganos, F., Sharpless, T. K., & Melamed, M. R. (1977). Cell cycle-related changes in nuclear chromatin of stimulated lymphocytes as measured by flow cytometry. Cancer Research, 37(12), 4635–4640.
- Greb, C. (2022). Fluorescent dyes. Leica Microsystems.
- Liu, K., Liu, P., Liu, R., & Wu, X. (2015). Dual AO/EB staining to detect apoptosis in osteosarcoma cells compared with flow cytometry. Medical Science Monitor Basic Research, 21, 15–20.
- Liu, M., Li, R., Tang, Y., Chang, J., Han, R., Zhang, S., Jiang, N., & Ma, F. (2017). New applications of the acridine orange fluorescence staining method: Screening for circulating tumor cells. Oncology Letters, 13(4), 2221–2229.
- Logos Biosystems. (2023). How to measure cell viability using Acridine Orange/Propidium Iodide: The principle and the procedure.
- Ma, T., Zhao, Y., & Cheng, X. (2024). Detection of dsRNA by Acridine Orange staining. Methods in Molecular Biology, 2771, 7–12.
- Murugan, S., & Amaravadi, R. K. (2016). Methods for studying autophagy within the tumor microenvironment. Advances in Experimental Medicine and Biology, 899, 145–166.
- Thomé, M. P., Filippi-Chiela, E. C., Villodre, E. S., Migliavaca, C. B., Onzi, G. R., Felipe, K. B., & Lenz, G. (2016). Ratiometric analysis of Acridine Orange staining in the study of acidic organelles and autophagy. Journal of Cell Science, 129(24), 4622–4632.
- Wikipedia contributors. (n.d.). Acridine orange. In Wikipedia.