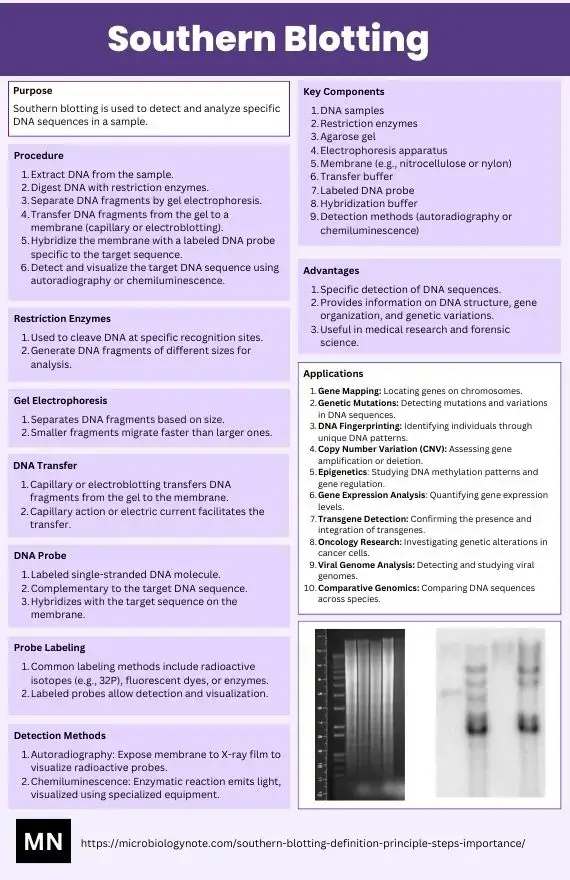
Southern blotting is a lab technique used to identify specific DNA sequences within a complex mixture. Developed by Edwin Southern in the 1970s, it starts by cutting DNA into fragments using enzymes that target precise locations. These fragments are then separated by size using gel electrophoresis, where an electric current pushes them through a gel matrix. Once sorted, the DNA is transferred from the gel onto a thin membrane—like moving ink from a page to a sheet of paper—creating a replica for easier handling.
Next, a probe designed to match the target DNA sequence is introduced. This probe, often tagged with a detectable marker (e.g., radioactive or fluorescent labels), binds exclusively to the matching fragments on the membrane. After washing away excess probe, the remaining signal reveals the presence and size of the target DNA.
Though newer methods have emerged, Southern blotting remains a foundational tool in genetics, forensics, and medical research. It’s handy for tasks like confirming gene mutations, analyzing genetic fingerprints, or verifying genetically modified organisms. While it might seem old-school compared to modern tech, its reliability keeps it in use for specific applications where precision matters.
What is Southern Blotting?
- In molecular biology, southern blotting is a method for specifically identifying certain DNA sequences within a DNA mixture.
- Edward M. Southern devised the method in 1975 and describes moving DNA fragments from an electrophoresisis gel to a membrane then hybridising with a labelled probe to find the target sequence.
- In DNA analysis, forensic science, and paternity testing, southern blotting is absolutely essential.
- Southern blotting generally consists in DNA digestion using restriction enzymes, gel electrophoresis, transfer to a membrane, hybridisation with a labelled probe, and fragment detection resulting from this hybridisation.
- Southern blotting finds genes rearranged, analyses DNA methylation patterns, detects mutations, and maps genes.
- The technique’s intricacy, need for high-quality DNA, and time-consuming character define some of its limits.
Southern Blotting Definition
Southern blotting is a molecular biology technique used to detect specific DNA sequences in a DNA sample by transferring and immobilizing the DNA fragments onto a solid membrane, followed by hybridization with a labeled DNA probe for detection.
Objectives
- Electrophoretic separation of DNA molecules by agarose gel electrophoresis
- Electrophoretic transfer of DNA from agarose gel to nylon membrane
- Immobilization of DNA on to nylon membrane
- Hybridization and non-isotopic detection of DNA of interest
- Teach the principle of Southern blotting.
- Explain principle of hybridization.
- To find the size of the DNA
Southern Blotting Principle
- The initial step involves the breakdown of DNA into smaller segments through the application of restriction enzymes.
- The fragments are subsequently separated by size using agarose gel electrophoresis.
- After electrophoresis, the DNA undergoes denaturation to form single strands, which are then transferred from the gel to a membrane.
- The membrane undergoes incubation with a labelled probe that exhibits complementarity to the target DNA sequence.
- The probe’s hybridisation with the target DNA facilitates the identification of particular DNA sequences.
- This method finds extensive application in gene mapping, detecting mutations, and conducting forensic analyses.
Requirements for Southern Blotting
- Necessary apparatus for the transfer and detection of DNA
- A water bath is utilised for accurate temperature control during the denaturation and hybridisation phases.
- Agarose gel apparatus, including the gel tray, combs, electrophoresis tank, and power supply, is utilised for the size-fractionation of DNA fragments.
- A UV transilluminator or UV crosslinker is utilised to visualise ethidium-bromide-stained DNA and to immobilise DNA on nylon membranes.
- Utilising a hybridisation oven or shaking incubator along with hybridisation bottles or trays is essential for achieving uniform probe binding.
- Imaging system or film processor designed for autoradiography or chemiluminescent detection of labelled probes
- Micropipettes along with compatible tips ensure precision in reagent handling.
- Microcentrifuge tubes utilised for the preparation of samples and conducting restriction digests
- Glass plates and Whatman 3 mm chromatography paper for the capillary transfer setup
- Nylon or nitrocellulose membranes serve as solid supports for the immobilisation of DNA.
- Syringe and cellulose acetate membrane (optional) for vacuum-blotting systems.
- Components and substances for fragment generation, labelling, and blocking
- Restriction endonucleases and their corresponding reaction buffers are utilised to cleave genomic or plasmid DNA into specific fragments.
- A glycerol-based dye mix serves as a DNA loading buffer to monitor sample migration throughout the electrophoresis process.
- A DNA labelling kit, whether radioactive or non-radioactive (such as digoxigenin or biotin labelling), is utilised for the preparation of complementary probes.
- Reagents for the detection of nucleic acids, such as autoradiography film and chemiluminescent substrates, are utilised for signal development.
- Utilising blocking agents like salmon or herring sperm DNA, Denhardt’s solution, or bovine serum albumin is essential to minimise non-specific probe binding.
- Sodium dodecyl sulphate (SDS) and formamide are utilised to enhance membrane prehybridization and manage stringency control.
- Polyvinylpyrrolidone and phenol (for specific non-radioactive detection methodologies)
- Compositions of solutions and buffer systems
- To prepare a denaturation buffer (1X), combine NaOH and NaCl in a 1:6 ratio, such as 0.5 M NaOH with 3 M NaCl.
- To prepare a neutralisation buffer at 1X concentration, mix Tris‑HCl and NaCl in a ratio of 5:3. For instance, use 0.5 M Tris‑HCl at pH 7.5 and 0.3 M NaCl.
- To prepare the transfer (blotting) buffer, start with a 20X SSC stock solution by dissolving 175.3 g of NaCl and 88.2 g of sodium citrate in 1 L of water. For capillary or vacuum transfer, dilute this solution to a 1X concentration.
- Electrophoresis buffer: 10X TBE—1.3 M Tris, 450 mM boric acid, 25 mM EDTA; utilise at 1X for agarose gel runs.
- Detection buffer (1X): A mixture of Tris‑HCl and NaCl in a 5:1 ratio (for instance, 0.5 M Tris‑HCl pH 7.5 and 0.1 M NaCl)
- Supplementary prehybridization buffer: 6X SSC, 5X Denhardt’s solution, 50% formamide, and 0.5% SDS to enhance probe binding and minimise background interference.
- Items for consumption and support
- Ethidium bromide or a different nucleic acid stain for confirming gel separation
- Utilise filter papers and paper towels to assemble the blotting “sandwich” and facilitate the wicking of transfer buffer.
- Weights or suction devices for capillary or vacuum blotting techniques
- X-ray film or digital imaging cassettes utilised for autoradiography detection
- Safety and quality assessments
- Proper radiation shielding and monitoring are essential when utilising radioactive probes.
- When working with caustic buffers such as NaOH and ethidium bromide, it is essential to wear nitrile gloves, a lab coat, and eye protection.
- DNA size markers are utilised to calibrate fragment sizes on gels and blots.
- Control groups that exhibit both favourable and unfavourable outcomes Samples of DNA are required to confirm the specificity of the probe and the stringency of hybridisation.
Preparation of Buffers
When preparing buffers for Southern blotting, it is important to use the correct concentrations of reagents to ensure optimal performance. Here are some commonly used buffers and their preparation methods:
- 10X TBE Buffer:
- Prepare a 1.3 M TRIS solution.
- Dissolve 450 mM boric acid in water.
- Add 25 mM EDTA to the solution.
- Adjust the pH if necessary.
- Mix the solutions thoroughly to obtain the 10X TBE buffer.
- 20X SSPE Buffer:
- Dissolve 2.98 M NaCl in water.
- Add 0.02 M EDTA to the solution.
- Prepare a 0.2 M phosphate buffer with a pH of 7.4.
- Mix the solutions together to obtain the 20X SSPE buffer.
- Denaturing Solution:
- Prepare a 1.5 M NaCl solution.
- Add 0.5 N NaOH to the solution.
- Adjust the pH to approximately 13.
- Neutralizing Solution:
- Dissolve 1.5 M NaCl in water.
- Prepare a 1 M TRIS HCl solution.
- Adjust the pH to 7.5.
- Denhardt’s Solution (50X):
- Dissolve 1% bovine serum albumin, 1% Ficoll, and 1% polyvinylpyrrolidone in water to a final volume of 50 mL.
- Sterilize the solution by filtration.
- 2X Prehybridization Solution:
- Combine 30 mL of 20X SSPE, 10 mL of 100X Denhardt’s solution, 10 mL of 10% SDS, and 50 mL of water to prepare a 1X prehybridization solution.
- Hybridization Solution:
- Mix 30 mL of 20X SSPE, 10 mL of 10% SDS, and 60 mL of water to prepare the hybridization solution.
- 1X Probe Buffer:
- Mix 50 µL of 1 M TRIS (pH 7.6), 5 µL of 2 M MgCl2, 10 µL of 0.5 M DTT, and 35 µL of water to prepare the probe buffer.
- 1X Probe Mix:
- Combine 2.7 µL of probe buffer, 2 µL of oligonucleotide probe (0.2 µg/µL), 1 µL of T4 phosphonucleotide kinase (PNK), 11.3 µL of water, and 10 µL of 32P-ATP to prepare the probe mix.
- 6X Low-Stringency Wash Solution:
- Mix 180 mL of 20X SSPE, 12 mL of 10% SDS, and 408 mL of water to prepare the low-stringency wash solution.
- 1X High-Stringency Wash Solution:
- Combine 30 mL of 20X SSPE, 12 mL of 10% SDS, and 558 mL of water to prepare the high-stringency wash solution.
By following these instructions, you can prepare the necessary buffers for Southern blotting, ensuring accurate and efficient detection of specific DNA sequences in your experiments.
| Buffer Name | Components | Preparation |
|---|---|---|
| 10X TBE Buffer | 1.3 M TRIS, 450 mM boric acid, 25 mM EDTA | Mix the reagents in the specified concentrations |
| 20X SSPE Buffer | 2.98 M NaCl, 0.02 M EDTA, 0.2 M phosphate | Combine the reagents in the specified amounts |
| Denaturing Solution | 1.5 M NaCl, 0.5 N NaOH | Mix the reagents and adjust the pH to ~13 |
| Neutralizing Solution | 1.5 M NaCl, 1 M TRIS HCl | Dissolve the reagents and adjust the pH to 7.5 |
| Denhardt’s Solution (50X) | 1% bovine serum albumin, 1% Ficoll, | Dissolve the reagents in water to a final volume |
| 1% polyvinylpyrrolidone | of 50 mL and sterilize by filtration | |
| 2X Prehybridization Solution | 30 mL 20X SSPE, 10 mL 100X Denhardt’s, | Mix the reagents in the specified proportions |
| 10 mL 10% SDS, 50 mL water | ||
| Hybridization Solution | 30 mL 20X SSPE, 10 mL 10% SDS, 60 mL water | Combine the reagents in the specified amounts |
| 1X Probe Buffer | 50 µL 1 M TRIS, 5 µL 2 M MgCl2, | Mix the reagents in the specified proportions |
| 10 µL 0.5 M DTT, 35 µL water | ||
| 1X Probe Mix | 2.7 µL probe buffer, 2 µL oligonucleotide | Combine the reagents in the specified amounts |
| probe (0.2 µg/µL), 1 µL T4 phosphonucleotide | ||
| kinase (PNK), 11.3 µL water, 10 µL 32P-ATP | ||
| 6X Low-Stringency Wash Solution | 180 mL 20X SSPE, 12 mL 10% SDS, 408 mL water | Mix the reagents in the specified proportions |
| 1X High-Stringency Wash Solution | 30 mL 20X SSPE, 12 mL 10% SDS, 558 mL water | Combine the reagents in the specified amounts |
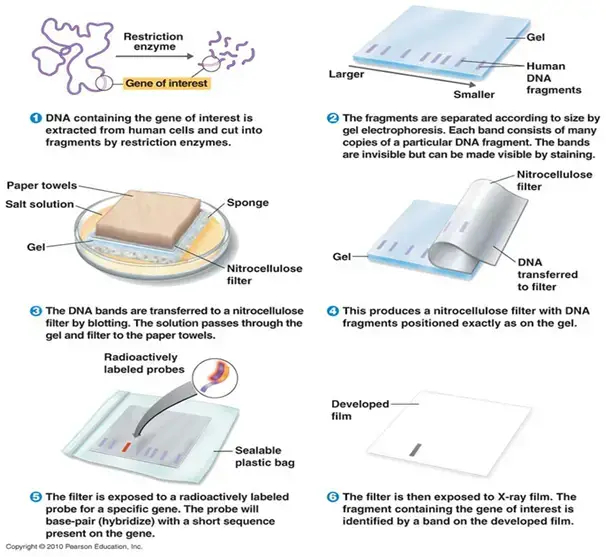
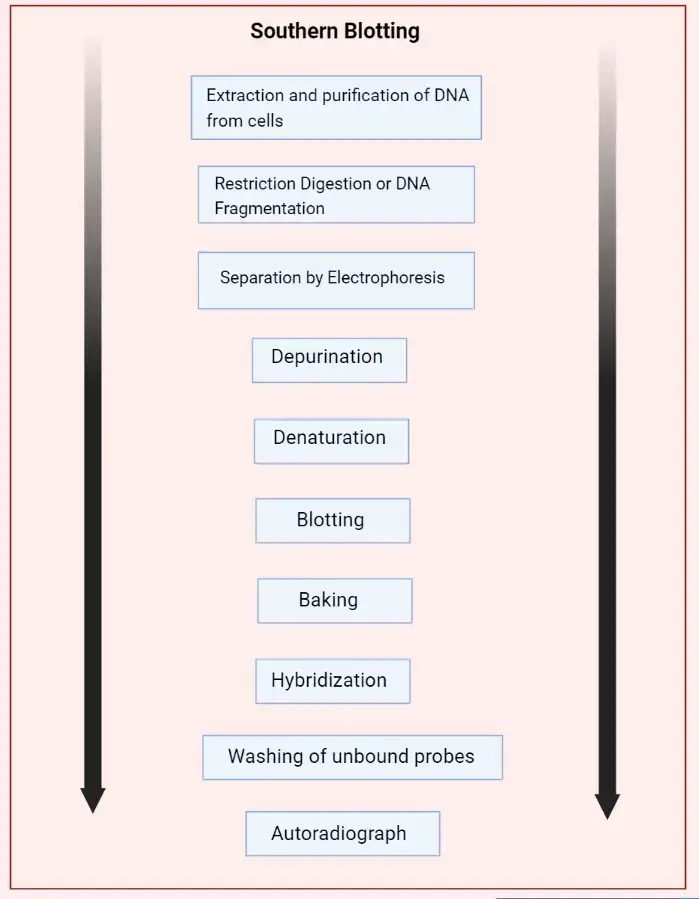
Basic Southern Blot Workflow
Southern blotting is a widely used technique in molecular biology for detecting specific DNA sequences within a sample. Here, we outline a basic workflow for performing a Southern blot, which can be adjusted based on specific experimental requirements.
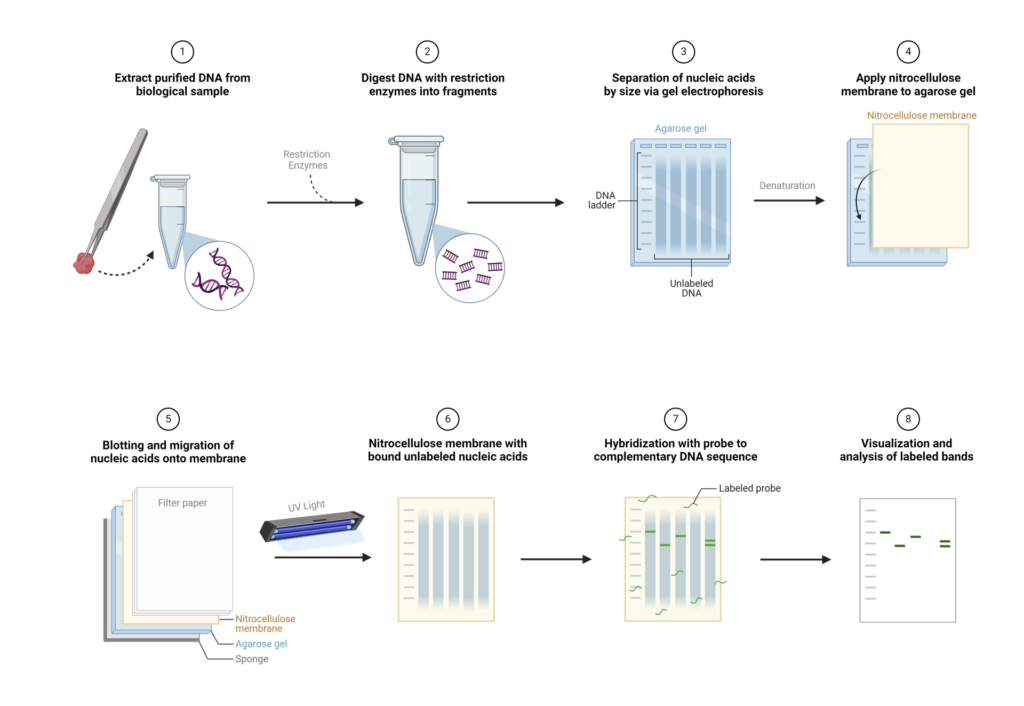
- Step 1: DNA Extraction – The process begins with the extraction of purified DNA from a biological sample, such as blood or tissue. The goal is to obtain high-quality, intact DNA suitable for subsequent analysis.
- Step 2: Restriction Digestion – Next, the extracted DNA is subjected to enzymatic digestion using one or more restriction enzymes. These enzymes cut the DNA at specific recognition sites, producing a mixture of fragments of varying lengths.
- Step 3: Gel Electrophoresis – The DNA fragments are then separated by gel electrophoresis. In this step, an electric current drives the DNA fragments through an agarose gel matrix. Because the gel acts like a sieve, smaller fragments migrate faster and farther than larger ones, resulting in a size-based separation.
- Step 4: Denaturation – Following electrophoresis, the DNA in the gel is denatured to convert the double-stranded DNA into single strands. This is typically achieved by soaking the gel in an alkaline solution, such as 0.5M NaOH. Only single-stranded DNA (ssDNA) can be transferred to the membrane in the subsequent step. For very large fragments (over 15kb), depurination can be performed by treating the gel with HCl to break the DNA into smaller fragments, which can then be neutralized to prepare for transfer.
- Step 5: Transfer to Membrane – The next step involves transferring the denatured DNA from the gel onto a solid support membrane, usually nylon due to its high binding capacity (approximately 500 µg/cm²), although nitrocellulose can also be used. This transfer can be done via capillary action or using a vacuum apparatus. The transfer process ensures that the DNA maintains its spatial arrangement from the gel onto the membrane.
- Step 6: DNA Fixation – Once the DNA has been transferred, it is covalently bound to the membrane using UV light or by baking at 80°C. This step immobilizes the DNA, preventing it from washing away during subsequent steps.
- Step 7: Hybridization – The membrane is then incubated with a labeled nucleic acid probe. This probe has a sequence complementary to the target DNA sequence and is tagged with a detectable marker, such as a fluorescent dye, radioactive isotope, or a chemiluminescent enzyme. During hybridization, the probe binds specifically to its complementary sequence on the membrane.
- Step 8: Washing – To remove any unhybridized probe, the membrane is washed with a buffer. This ensures that only the probe molecules that have hybridized to their target DNA remain on the membrane.
- Step 9: Detection – The hybridized probes are then detected and visualized using methods appropriate for the label used. For instance, radiolabeled probes can be visualized using X-ray film or phosphorimaging, while probes labeled with chemiluminescent enzymes are detected using substrates that produce light in the presence of the enzyme.
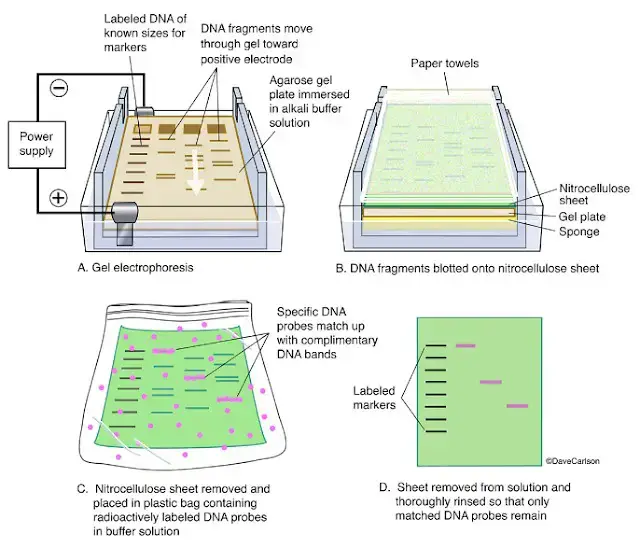
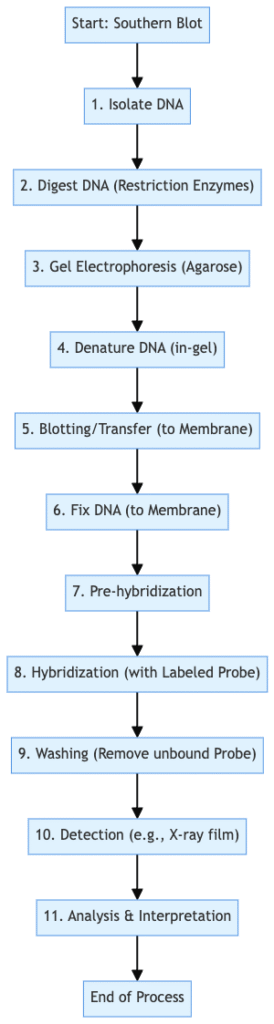
Southern Blotting Procedure
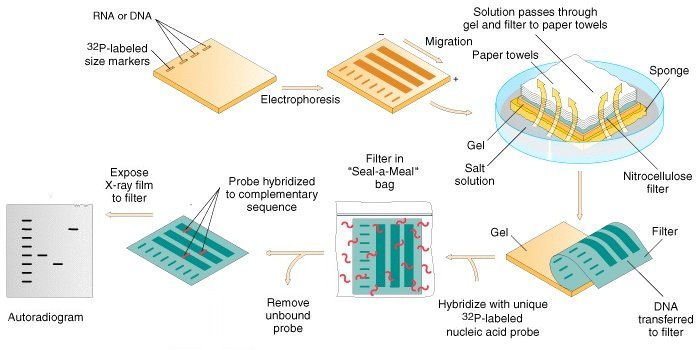
- Restriction Digestion of DNA
- About 10 µg of genomic DNA is digested with the appropriate restriction enzyme in a microcentrifuge tube.
- Incubate the tube overnight at 37°C.
- Optionally, heat at 65°C for 20 minutes after incubation to denature the restriction enzymes.
- Add 10 µl of DNA sample buffer, then load the mixture onto an agarose gel for electrophoresis.
- Electrophoresis
- Choose the gel percentage based on the DNA fragment size to be separated and prepare the gel.
- Prepare electrophoresis buffer by adding ethidium bromide and pour it into the electrophoresis tank, ensuring it is a few millimeters above the gel.
- Set up a gel cast with a comb to form wells for the sample loading. Pour the gel and remove the comb once it has solidified.
- Place the gel into the tank and cover it with running buffer.
- Prepare DNA samples by adding loading buffer, and carefully pipette the samples into the wells.
- Connect the power supply and run the gel overnight.
- Denaturation
- After electrophoresis, transfer the gel into a glass tray containing 500 ml denaturation buffer (1.5 M NaCl and 0.5 M NaOH) for 45 minutes at room temperature.
- Replace the denaturation buffer with neutralization buffer, then allow the gel to soak for 1 hour with slow rotation.
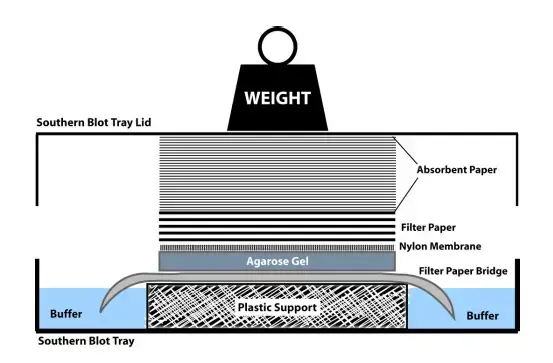
- Blotting
- Prepare a glass dish with an oblong sponge larger than the gel, partially submerged in SSC buffer.
- Cut three pieces of Whatman 3 mm paper to the same size as the sponge, place them on the sponge, and wet with SSC.
- Place the gel on top of the filter paper and remove air bubbles by gently rolling a glass pipette over the surface.
- Position a nylon membrane large enough to cover the gel on top of the gel and flood with SSC.
- Add a few sheets of filter paper on top of the membrane and place a glass plate to hold everything in place.
- Allow the DNA transfer to occur overnight.
- Baking/Immobilization
- Remove the nylon membrane from the blotting structure and bake at 80°C for 2-3 hours in a vacuum or regular oven.
- Alternatively, DNA strands on the membrane can be immobilized by exposing it to ultraviolet radiation.
- Hybridization
- Expose the membrane to a hybridization probe (DNA or RNA) that specifically binds to the target DNA sequence.
- The probe is labeled with a detectable marker (radioactive, fluorescent, or chromogenic).
- Conditions are optimized for probe hybridization with the complementary target sequence on the membrane.
- After hybridization, wash the membrane with a buffer to remove nonspecific binding, leaving only the labeled probe bound to the target sequence.
- Detection
- Detect the hybridized DNA regions using autoradiography by placing the nylon membrane in contact with photographic film.
- The film reveals the position of the hybridized DNA, helping determine the length of the fragments by comparing them with marker DNA of known size.
- Alternatively, use fluorescent or chromogenic dyes to visualize the hybridized fragments, either through X-ray film or by color development on the membrane.

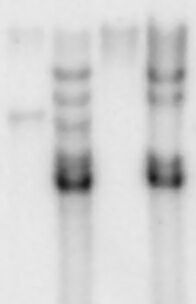
Observation
Observe for a single blue band on the nylon membrane.
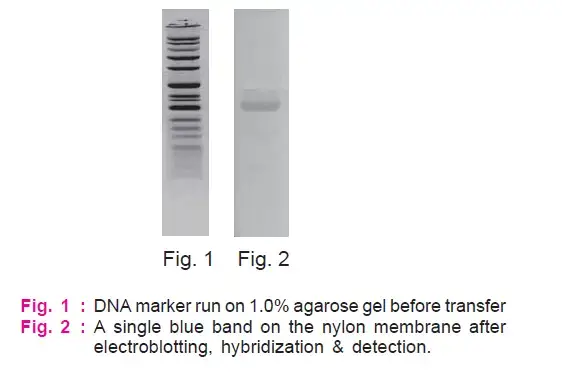
Precautions
- Always wear non-powdered gloves and a lab coat to ensure sample integrity and safeguard against hazardous or radioactive substances.
- Utilise exclusively DNase- and RNase-free glassware, plasticware, buffers, and water to prevent the degradation of target DNA.
- Utilise filter-barrier pipette tips and ensure to change tips between each sample or reagent to avoid cross-contamination.
- Establish and clearly delineate “nuclease‑free” workspaces and tools specifically for pre‑ and post‑hybridization procedures.
- Pre-wet the Whatman paper, membrane, and sponge in transfer buffer to ensure uniform capillary flow and avoid dry spots.
- Eliminate any air bubbles beneath the gel and membrane by gently rolling a glass rod over the surface to guarantee uniform DNA transfer.
- Carefully observe the agarose melting process during heating to avoid overheating or boiling over, as this can compromise DNA integrity and alter the gel structure.
- Carefully detach the gel comb to prevent the collapse of wells and ensure the integrity of the samples is maintained.
- Once the membrane is positioned, it is crucial to avoid shifting or disturbing it during the transfer setup to ensure the precise alignment of DNA bands is maintained.
- Ensure that line work surfaces are equipped with absorbent bench pads and utilise suitable shielding, such as Plexiglas, when managing radioactive probes.
- Conduct a survey of the work area and personnel using a Geiger counter prior to and following exposure to ³²P probes to ensure radiation safety compliance.
- Membranes should be baked at 80 °C for a duration of 2–3 h, or alternatively, UV-crosslinking should be performed at the recommended energy level (e.g., 120 mJ/cm²) immediately following transfer to ensure the immobilisation of DNA.
- Utilise positively charged nylon membranes along with a high-salt (20X SSC) buffer for transfer to enhance DNA binding and reduce background interference.
Application of Southern blotting
- Detection of a single gene within complex DNA samples
- Gene mapping and RFLP marker analysis to locate genes on chromosomes
- Identification of chromosomal or gene rearrangements, insertions, deletions, and point mutations in DNA
- Restriction mapping and SNP analysis to determine altered restriction sites
- Homology‑based screening of DNA libraries for gene cloning
- Comparative genomics to find homologous sequences across species
- DNA fingerprinting for forensic identification and paternity testing
- Diagnosis of genetic disorders such as sickle cell anemia and monoclonal leukemia
- Analysis of gene copy number changes in cancer cells to detect amplifications and deletions
- Epigenetic studies by assessing DNA methylation status with methylation‑sensitive restriction enzymes
- Verification of transgene integration and copy number in genetically modified organisms
- Clinical diagnosis of triplet repeat expansion disorders (e.g., myotonic dystrophy type 1)
- Pathogen strain typing for epidemiological outbreak investigation
Limitations of Southern Blot
- Demands considerable amounts (5–10 µg) of high-quality, intact DNA, which can pose challenges when sourcing from limited or degraded samples.
- The workflow is quite labor-intensive, involving digestion, electrophoresis, transfer, hybridization, and detection, which typically takes 2 to 3 days to complete.
- Characterized by significant labor demands and intricate technical requirements, this process involves numerous manual steps that require skilled individuals to minimize the risk of mistakes.
- The limited throughput allows for the processing of only a small number of samples per blot, rendering it inappropriate for extensive studies.
- Semi-quantitative analysis indicates that band intensities yield approximate rather than absolute measures of DNA abundance.
- The elevated expenses arise from the costly enzymes, labeled probes (whether radioactive or non-radioactive), membranes, and imaging supplies.
- Insufficient restriction digestion or ineffective transfer may result in ambiguous or absent bands.
- Conventional capillary transfer techniques can lead to the compression of gels due to weight, which diminishes transfer efficiency.
- The resolution is constrained for extremely small fragments or single-base alterations; detection of point mutations necessitates supplementary methods.
- Primarily replaced by PCR-based techniques for various applications, yet still utilized in scenarios where PCR could introduce bias (e.g., gene rearrangement analysis).
Troubleshooting of Southern blotting
Troubleshooting is an important aspect of Southern blotting to identify and resolve issues that may arise during the process. Here are some common troubleshooting steps to consider:
- Leaking or Floating Samples:
- Samples may leak out of the wells if there is damage or puncture. Be cautious when removing the comb and loading the gel.
- Floating of samples can occur due to residual ethanol or hasty gel loading. Ensure ethanol is completely removed and load the gel slowly.
- Frowned Appearance of Bands: If the bands on the gel have a frowned appearance, it may be due to running the gel at a high voltage. Choose a lower voltage to resolve this issue.
- Invisibility of DNA and Molecular Weight Markers: If DNA, including the molecular weight markers, is not visible, it could be due to using a low amount of ethidium bromide. Adjust the concentration of ethidium bromide appropriately.
- Non-separation of DNA and Molecular Weight Markers: If the DNA and molecular weight markers are not separated, it may be because the gel is made up of water instead of 1X running buffer. Ensure the gel is prepared using the correct running buffer.
- DNA Sticking to Wells: When the DNA concentration in the sample is too high, it can cause the DNA to stick to the wells. Reduce the DNA concentration in the sample to prevent this issue.
- Background Spots: Appearance of background spots could be due to powder from gloves. Use powder-free gloves to eliminate this problem. Increasing the number of washes can also help remove the background.
- Low Signal Intensity:
- If the signal intensity is low, it may be due to a low sample content. Increase the DNA concentration in the sample to improve the signal.
- Additionally, check the transfer time and hybridization timing. A shorter transfer time or inadequate hybridization can affect the signal intensity. Optimize the transfer time and hybridization conditions accordingly.
Southern Blotting Concept map
FAQ
What is Southern blotting?
Southern blotting is a laboratory technique used to detect specific DNA sequences in a sample. It involves the separation of DNA fragments by gel electrophoresis, transfer of the fragments to a membrane, and hybridization with a labeled probe to visualize the target sequence.
What is the purpose of Southern blotting?
Southern blotting is primarily used to detect and analyze specific DNA sequences in a sample. It can be employed for various applications, such as gene mapping, identification of genetic mutations, DNA fingerprinting, and studying gene expression patterns.
How does Southern blotting work?
Southern blotting involves several steps. First, DNA is extracted from the sample and digested with restriction enzymes to produce DNA fragments. These fragments are then separated by size using gel electrophoresis. After that, the DNA fragments are transferred to a membrane, where they are immobilized. Finally, the membrane is exposed to a labeled DNA probe that hybridizes specifically to the target DNA sequence, allowing detection and visualization of the sequence of interest.
What are the key components required for Southern blotting?
The key components for Southern blotting include DNA samples, restriction enzymes, agarose gel, electrophoresis apparatus, a membrane (such as nitrocellulose or nylon), transfer buffer, labeled DNA probe, hybridization buffer, and detection methods (such as autoradiography or chemiluminescence).
What is the role of restriction enzymes in Southern blotting?
Restriction enzymes are used in Southern blotting to cleave DNA at specific recognition sites. By digesting the DNA sample with appropriate restriction enzymes, it is possible to generate fragments of different sizes, allowing the identification and analysis of specific DNA sequences.
How is the DNA transferred from the gel to the membrane during Southern blotting?
The DNA fragments are transferred from the gel to the membrane through a process called capillary or electroblotting. The gel is placed on top of the membrane, and a buffer solution is allowed to flow through the gel by capillary action or through the application of an electric current. This transfers the DNA fragments onto the membrane, where they become immobilized.
What is the purpose of the DNA probe in Southern blotting?
The DNA probe is a labeled single-stranded DNA molecule that is complementary to the target DNA sequence of interest. It is used to specifically hybridize with the target sequence on the membrane after the transfer step. The labeled probe allows the detection and visualization of the target DNA sequence.
How is the DNA probe labeled in Southern blotting?
The DNA probe can be labeled with a variety of markers, such as radioactive isotopes (e.g., 32P), fluorescent dyes, or enzymes. These markers allow the visualization and detection of the probe after hybridization with the target DNA sequence.
What are the detection methods used in Southern blotting?
The two common methods of detecting the DNA probe in Southern blotting are autoradiography and chemiluminescence. Autoradiography involves exposing the membrane to X-ray film, while chemiluminescence relies on the emission of light from an enzymatic reaction between the probe and a substrate.
What are the advantages of Southern blotting?
Southern blotting is a versatile technique that allows the specific detection of DNA sequences. It provides valuable information about DNA structure, gene organization, and genetic variations. It can also be used for diagnostic purposes in medical research and forensic science. However, it is a time-consuming and labor-intensive technique and has been largely replaced by more advanced methods like PCR and DNA sequencing for routine DNA analysis.
- Mellars., & Gomez., k. (2011). Mutation detection by Southern blotting. Methods Mol Biol, 688, 281-291.
- Brown, T. (2001). Southern blotting. Curr Protoc Immunol, Chapter 10:Unit 10.
J. Aubin., H. Collandre., D. Candotti., D. Ingrand., C. Rouzioux., M. Burgard., . . . Agut., H. (1991). - Several groups among human herpesvirus 6 strains can be distinguished by Southern blotting and polymerase chain reaction. J Clin Microbiol, 29(2), 367-72.
- C. Lo., M. Coulling., & Kirby., C. (1987). Tracking of mouse cell lineage using microinjected DNA sequences: analyses using genomic Southern blotting and tissue-section in situ hybridizations. Differentiation, 35(1), 37-44.
- Ohshima., M. Kikuchi., F. Eguchi., Y. Masuda., Y. Sumiyoshi., H. Mohtai., . . . Kimura., N. (1990). Analysis of Epstein-Barr viral genomes in lymphoid malignancy using Southern blotting, polymerase chain reaction and in situ hybridization. Virchows Arch B Cell Pathol Incl Mol Pathol, 59(6), 383-90.
- Gaurab Karki, Southern Blotting: principle, procedure and application, Published on December 3, 2017, Online Biology Notes. https://www.onlinebiologynotes.com/southern-blotting-principle-procedure-application/
- https://www.sigmaaldrich.com/technical-documents/articles/biology/southern-and-northern-blotting.html
- https://www.mybiosource.com/learn/southern-blotting/
- https://laboratoryinfo.com/southern-blot/
- https://geneilabs.com/product/genei-southern-hybridization-teaching-kit-with-ets5-5-expts/