What is Sanger Sequencing?
- Sanger sequencing, also known as dideoxy sequencing or chain termination method, identifies the order of nucleotide bases in DNA. This method relies on chain termination by modified nucleotides called dideoxynucleotide triphosphates (ddNTPs). Developed in 1977 by Frederick Sanger and colleagues, it was the first DNA sequencing method and remains highly regarded for its accuracy and the length of the reads it produces.
- Over the years, sequencing technologies have evolved significantly. Despite these advancements, Sanger sequencing remains widely used, especially for small-scale projects. Its precision and reliability make it a valuable tool in various research applications. Next-generation sequencing (NGS) technologies, such as Illumina, PacBio, and Nanopore, have emerged for higher-throughput sequencing projects. These technologies can sequence larger amounts of DNA more quickly and cost-effectively. Nevertheless, Sanger sequencing still holds a niche, particularly for validating NGS results and sequencing small, targeted regions of the genome.
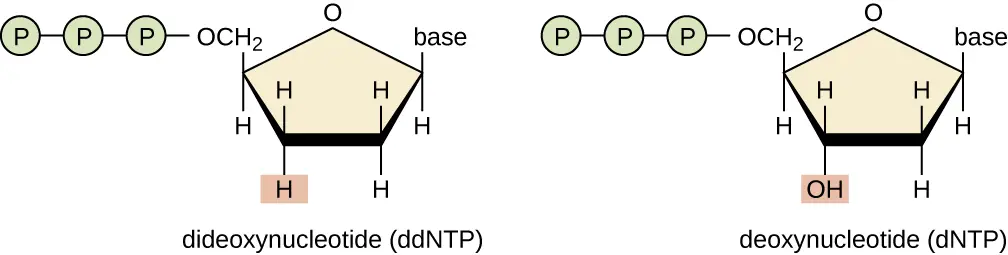
- The technique involves the incorporation of ddNTPs into a growing DNA chain during replication. These ddNTPs lack a 3′ hydroxyl group, which prevents the addition of further nucleotides, thus terminating the chain. By incorporating fluorescent or radioactive labels, the terminated fragments can be detected and analyzed to determine the DNA sequence.
- Sanger sequencing was crucial for the Human Genome Project, completed in 2003, marking a significant milestone in genomics. Today, it remains a fast and cost-effective method for reading small, targeted regions of the genome. Its applications include testing for known familial variants, validating NGS results, and single gene sequencing.
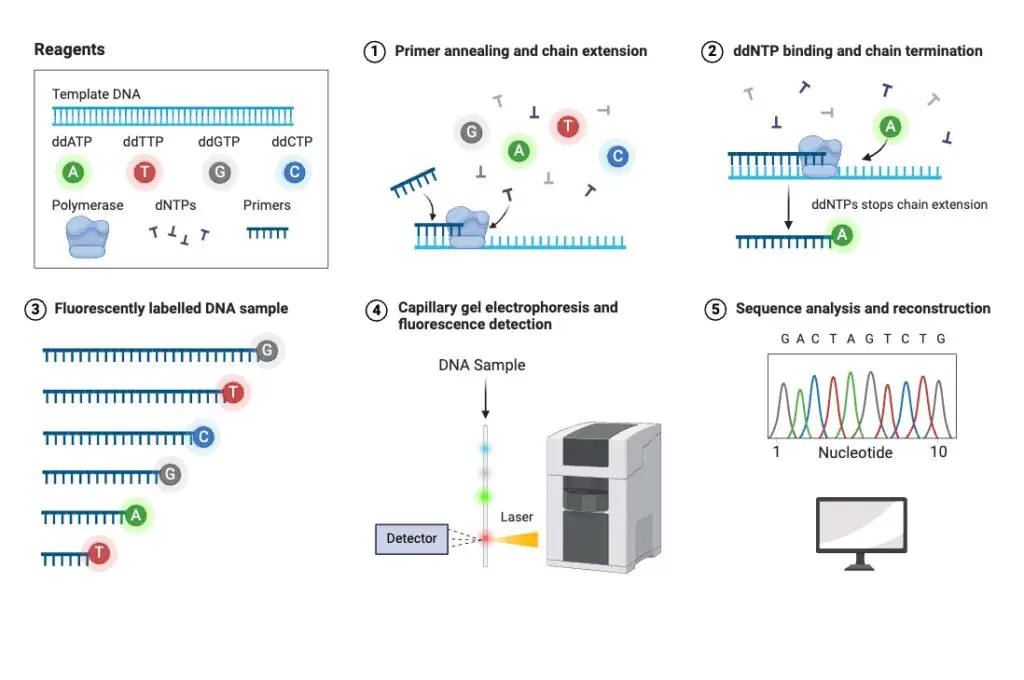
Principle of Sanger Sequencing
The principle of Sanger sequencing is rooted in the termination of DNA strand elongation by dideoxynucleotide triphosphates (ddNTPs). These ddNTPs are modified versions of normal DNA nucleotides that lack a 3’ hydroxyl group, which is essential for forming a phosphodiester bond. Without this bond, the DNA strand cannot elongate further.
In the sequencing process, a mix of labeled ddNTPs, regular deoxyribonucleotide triphosphates (dNTPs), and the template DNA is used in a Polymerase Chain Reaction (PCR). The DNA polymerase enzyme adds dNTPs to the growing DNA strand. Occasionally, a ddNTP is incorporated instead of a dNTP. When this happens, the chain elongation stops because the ddNTP lacks the 3’ hydroxyl group needed for the next nucleotide to attach.
The result is a collection of DNA fragments of varying lengths. Each fragment ends with a labeled ddNTP. These fragments are then separated by electrophoresis, which arranges them by size. The labels on the ddNTPs are fluorescent, allowing detection by a laser or an imaging system. By analyzing the fluorescent signals, the sequence of the DNA can be determined. The different colored labels correspond to the four bases (adenine, thymine, cytosine, and guanine), revealing the order in which they appear in the DNA sequence.
Therefore, Sanger sequencing uses ddNTPs to create a series of DNA fragments terminated at each occurrence of a specific base. The sequence of these fragments is then read to determine the original DNA sequence. This method, despite being supplemented by more advanced technologies, remains a cornerstone of genetic research due to its high accuracy and reliability.
How does Sanger sequencing work?
Sanger sequencing is a methodical process that accurately determines the sequence of DNA. Here’s a detailed explanation of how it works:
- Preparation of Patient DNA
- The process begins with extracting the patient’s DNA, which will serve as the template for sequencing. The DNA is then denatured into single strands.
- Polymerase Chain Reaction (PCR)
- The denatured DNA is subjected to PCR. During PCR, the DNA is mixed with a combination of normal nucleotides (dNTPs) and chain-terminating nucleotides (ddNTPs). Each ddNTP is labeled with a distinct fluorescent marker.
- Incorporation of ddNTPs
- The DNA polymerase enzyme synthesizes new DNA strands by adding dNTPs. Occasionally, a ddNTP is incorporated instead of a dNTP. When this happens, the chain elongation stops because ddNTPs lack the 3’ hydroxyl group necessary for the addition of further nucleotides. This results in DNA fragments of varying lengths, each ending with a fluorescently labeled ddNTP.
- Separation by Capillary Electrophoresis
- The mixture of DNA fragments is then subjected to capillary electrophoresis. This technique separates the DNA fragments by size. Shorter fragments travel faster through the capillary tube, while longer fragments move more slowly.
- Detection of Fluorescent Labels
- As the fragments pass through a detector, a laser excites the fluorescent labels on the ddNTPs. The detector identifies the specific fluorescent signal of each ddNTP, indicating the terminating base of each fragment.
- Generation of a Chromatogram
- The sequence data is then compiled into a chromatogram. This graphical representation shows the order of the bases in the DNA fragment. Each peak in the chromatogram corresponds to a nucleotide, and the sequence is read from the shortest to the longest fragment.
- Comparison to Reference Sequence
- The obtained sequence is compared to a reference sequence to identify any genetic variants. This comparison helps in detecting mutations or alterations in the DNA that may be associated with specific diseases or conditions.
Sanger sequencing’s methodical approach, combining PCR, selective termination, and precise separation, provides high accuracy and reliability, making it invaluable in genetic research and clinical diagnostics.
Sanger Sequencing Steps
Sanger sequencing involves four key steps. These steps ensure accurate determination of the DNA sequence.
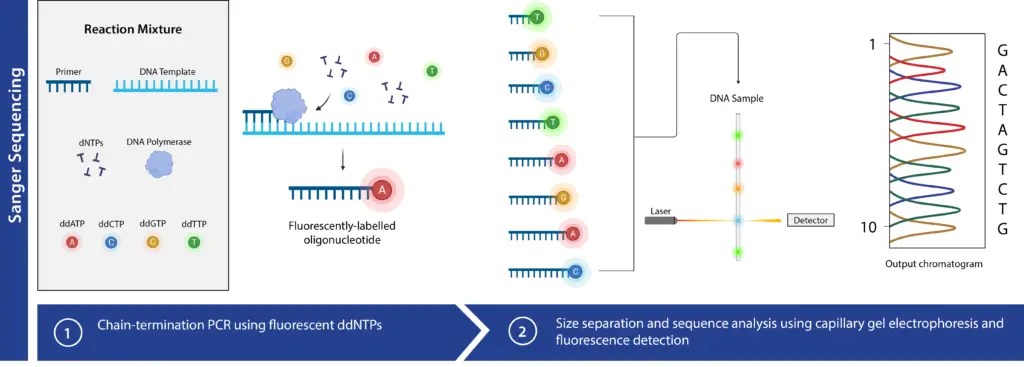
- DNA Template Preparation
- The first step is extracting the DNA of interest. DNA can be extracted using methods like chemical extraction, column-based extraction, or magnetic bead-based extraction.
- Identical, single-stranded DNA molecules are prepared for sequencing. This preparation is crucial for the accuracy and efficiency of the sequencing process.
- Chain Termination PCR
- The target DNA is amplified using Polymerase Chain Reaction (PCR). This process includes initial denaturation, followed by cycles of denaturation, annealing, and extension, and concludes with a final hold at 4°C.
- The PCR mix contains template DNA, primers, all four deoxyribonucleotide triphosphates (dNTPs), and DNA polymerase. Importantly, small amounts of dideoxynucleotide triphosphates (ddNTPs), each labeled with a distinct fluorescent marker, are added.
- ddNTPs lack the 3’ hydroxyl group, causing chain termination when incorporated. Due to their lower concentration compared to dNTPs, chain termination occurs at various points, producing a collection of DNA strands of different lengths, each ending in a ddNTP.
- Traditionally, this reaction is carried out in four separate tubes, each containing one type of labeled ddNTP. In automated Sanger sequencing, all four ddNTPs are included in a single reaction, each with a unique fluorescent label.
- Before electrophoresis, a sequencing clean-up step removes unincorporated ddNTPs and other contaminants.
- Separation of DNA Fragments
- Electrophoresis is used to separate the DNA fragments by length. This can be performed using polyacrylamide gel electrophoresis or capillary gel electrophoresis.
- Traditional polyacrylamide gel electrophoresis separates fragments by size, with each PCR reaction run in separate lanes to identify the corresponding ddNTP.
- Capillary electrophoresis (CE) employs glass capillaries filled with a gel polymer. Each sequencing reaction runs in a single capillary, enhancing automation and efficiency.
- Detection and Analysis
- After separation, DNA fragments pass through a fluorescence detector. This detector identifies the fluorescent label on the ddNTP at the end of each fragment.
- Each fluorescent signal represents a specific nucleotide, allowing the generation of a chromatograph that shows the peaks of each labeled fragment.
- By reading the order of the fluorescent labels, the sequence of the DNA template is determined accurately.
Advantages of Sanger Sequencing
Sanger sequencing remains the gold standard method for many research and clinical applications due to several distinct advantages.
- High Accuracy
- Sanger sequencing provides highly accurate results, making it ideal for validating sequences obtained through next-generation sequencing (NGS) technologies. This accuracy is crucial for confirming single nucleotide variants and small insertions or deletions.
- Well-Established Technology
- The technology is well-established and involves a straightforward process, leading to reliable and reproducible outcomes. Researchers and clinicians have confidence in the robustness of this method.
- Suitable for Small-Scale Projects
- It is particularly suitable for small-scale projects or those involving short DNA regions. Sanger sequencing excels in sequencing small, targeted areas, such as single genes or specific variants.
- Long Read Lengths
- Sanger sequencing produces relatively long read lengths, which can reach up to approximately 1000 base pairs (bp). This is advantageous for obtaining detailed sequence information over longer stretches of DNA compared to the shorter reads typical of many NGS platforms.
- Simplified Data Analysis
- Data analysis from Sanger sequencing is straightforward and does not require the complex bioinformatics tools and expertise often necessary for interpreting NGS data. The chromatograms generated are clear and easily interpretable.
- Cost-Effectiveness
- For urgent testing where single samples cannot be batched, Sanger sequencing is cost-effective. This is particularly useful in scenarios such as prenatal testing or parental carrier testing during pregnancy, where timely results are critical.
- Flexibility
- Sanger sequencing offers flexibility for testing specific familial variants. It allows for focused analysis without the need for broad, high-throughput sequencing.
- High-Quality Data
- The quality of the data obtained from Sanger sequencing is high, providing clear and precise results. This quality is essential for applications that demand exact sequence determination.
Limitations of Sanger Sequencing
Sanger sequencing, while a foundational technique in molecular biology, has several limitations. These constraints highlight why newer methods have been developed for broader applications.
- Limited Throughput
- Sanger sequencing can only sequence short DNA fragments and processes one fragment at a time. This makes it slow and inefficient for sequencing large genomes or numerous samples simultaneously.
- Cost Inefficiency
- The method is not cost-effective for sequencing many genes in parallel or for repeated sequencing of the same region across multiple samples. High costs and low throughput make it unsuitable for large-scale projects.
- Detection Limitations
- Sanger sequencing may not detect mosaicism effectively. Mosaicism involves the presence of multiple genetic cell lines within the same individual, which can be missed due to the technique’s sensitivity limits.
- Sample Requirements
- This method often requires a larger amount of input DNA compared to next-generation sequencing (NGS). It also demands high-quality, pure DNA samples, as degraded or contaminated DNA can produce unreliable results.
- Fragment Length Restrictions
- Sanger sequencing is restricted to short DNA sequences. Its capacity to read longer sequences is limited, making it less useful for comprehensive genomic studies.
- Manual Processing
- The technique involves more manual steps compared to automated NGS workflows, resulting in longer preparation and handling times. This manual aspect increases the complexity and time required for sequencing.
- Low Sensitivity
- The method has low sensitivity, which hinders the detection of rare mutations and the analysis of complex mixtures. This limitation is significant when studying genetic heterogeneity within a sample.
- Time-Consuming
- Sanger sequencing is a time-consuming process. The sequencing, data collection, and analysis phases take longer compared to the rapid workflows of modern NGS techniques.
Applications of Sanger Sequencing
Sanger sequencing remains a crucial tool in various scientific and clinical fields due to its accuracy and reliability. Here are the primary applications:
- Medical Diagnosis
- Sanger sequencing is extensively used to identify genes associated with various diseases. It is especially valuable for targeted sequencing of specific genomic regions, aiding in clinical diagnostics and genetic testing. Therefore, it helps in diagnosing hereditary conditions and validating results from high-throughput sequencing methods.
- Species Identification and Evolutionary Studies
- The technique is pivotal in identifying new species by comparing their gene sequences with known species. Besides, it helps in studying the evolutionary history and genetic diversity of different organisms. This application is essential in taxonomy and phylogenetics.
- Forensic Science
- In forensic science, Sanger sequencing is employed for personal identification and DNA fingerprinting. It is instrumental in analyzing DNA evidence in criminal cases, thereby contributing to the justice system by providing accurate genetic data for investigations.
- Agriculture
- Sanger sequencing plays a significant role in agriculture by identifying different breeds of crops and livestock. It is also used in the conservation of endangered species. Therefore, it aids in improving crop varieties and breeding programs through precise genetic analysis.
- Newer Technologies
- The method finds applications in cutting-edge technologies such as single-cell sequencing and synthetic biology. In single-cell sequencing, it helps understand the genetic makeup of individual cells. In synthetic biology, Sanger sequencing verifies the accuracy of synthetic DNA constructs.
Clinical applications of Sanger sequencing
Sanger sequencing remains a cornerstone in clinical genomics due to its accuracy and reliability. It is utilized in several key applications within clinical laboratories:
- Diagnostic Sequencing of Single Genes
- Sanger sequencing is often employed for the precise diagnosis of single-gene disorders. It enables the direct examination of specific genes to identify pathogenic mutations responsible for genetic conditions.
- Testing for Familial Sequence Variants
- Predictive Genomic Testing: This includes testing at-risk relatives for known familial variants, such as BRCA1 mutations associated with breast cancer. It helps predict the likelihood of developing hereditary diseases.
- Carrier Testing: Used to determine if parents carry genetic mutations that could lead to autosomal recessive conditions in their offspring. For instance, testing for cystic fibrosis carriers can inform reproductive decisions.
- Prenatal Testing: Sanger sequencing can be applied to test for known familial genetic variants in prenatal samples, aiding in the early detection of genetic disorders before birth.
- Segregation Analysis: This analysis is crucial for understanding the pathogenicity of a variant. It involves testing family members to see if a genetic variant is present in affected relatives and not in unaffected ones.
- Confirmation of Variants Identified by Next-Generation Sequencing (NGS)
- Sanger sequencing is frequently used to confirm variants detected by NGS technologies. Due to its high accuracy, it serves as a reliable method for validating NGS results.
- Filling Gaps in NGS Data
- In some cases, NGS data may be incomplete or ambiguous. Sanger sequencing is employed to address these gaps, ensuring comprehensive and accurate genetic information.
References
- Zhang, Lu & Chen, FengXin & Zeng, Zhan & Xu, Mengjiao & Sun, Fangfang & Yang, Liu & Bi, Xiaoyue & Lin, Yanjie & Gao, YuanJiao & Hao, HongXiao & Yi, Wei & Li, Minghui & Xie, Yao. (2021). Advances in Metagenomics and Its Application in Environmental Microorganisms. Frontiers in Microbiology. 12. 10.3389/fmicb.2021.766364.
- Gauthier, Michel. (2007). Simulation of polymer translocation through small channels: A molecular dynamics study and a new Monte Carlo approach.
- https://www.aatbio.com/catalog/sanger-sequencing
- https://www.genomicseducation.hee.nhs.uk/genotes/knowledge-hub/sanger-sequencing/
- https://www.boundless.com/microbiology/textbooks/boundless-microbiology-textbook/microbial-genetics-7/bioinformatics-83/dna-sequencing-based-on-sanger-dideoxynucleotides-459-6637/
- http://www.hhmi.org/biointeractive/sanger-method-dna-sequencing
- https://www.khanacademy.org/science/biology/biotech-dna-technology/dna-sequencing-pcr-electrophoresis/a/dna-sequencing
- https://www.dnalc.org/view/15479-Sanger-method-of-DNA-sequencing-3D-animation-with-narration.html
- https://www.thermofisher.com/np/en/home/life-science/sequencing/sanger-sequencing/sanger_sequencing_method.html
- http://www.ithanet.eu/ithapedia/index.php/Protocol:DNA_sequencing