What is restriction digestion?
In molecular biology restriction digestion is used to prepare DNA for analysis or other processing. It is also known as DNA fragmentation. According to Hartl and Jones Restriction Digestion is a;
This enzymatic technique can be used for cleaving DNA molecules at specific sites, ensuring that all DNA fragments that contain a particular sequence at a particular location have the same size; furthermore, each fragment that contains the desired sequence has the sequence located at exactly the same position within the fragment. The cleavage method makes use of an important class of DNA-cleaving enzymes isolated primarily from bacteria. These enzymes are called restriction endonucleases or restriction enzymes, and they are able to cleave DNA molecules at the positions at which particular short sequences of bases are present.
- In 1970, Hamilton-Smith published a paper on the discovery and purification of the first restriction enzyme, or endonuclease, HindII; for which he was awarded the Noble Prize in medicine.
- These enzymes are invaluable in molecular biology, cloning, genetic engineering, and multiple other scientific disciplines
- The restriction enzymes cleave the DNA molecule at a specific site or specific nucleotide sequence, is known as recognition sequences or Restriction sites.
- What are possible applications for restriction digestion? well, restriction digest is used in different analytical techniques such as, Restriction fragment length polymorphism (RFLP), Short tandem repeat polymorphism (AFLP), Amplified fragment length polymorphism (STRP).
Overview
- To prevent the attack of bacteriophage, bacteria develop different enzymes which are known as endonucleases that chop up any foreign DNA.
- Hence this enzyme restricts the bacteriophage infection, they are also termed “restriction endonucleases”.
- In molecular biology restriction endonucleases are also known as molecular scissors due to their ability to cut the DNA into fragments.
- These enzymes are mainly found in the cytoplasm of bacterial cell, hence the bacteria protects their DNA by methylating the adenine or cytosine bases. These methyl groups prevent the binding of restriction enzymes, but not the normal reading and replication of genetic information. While the bacteriophage DNA lacks these protective methyl groups and will be destroyed.
- The restriction enzyme with the modification methyltransferase together develops a restriction modification (RM) system. There are mainly four types of RM systems which are distinguished based on subunit composition, kinds of sequences recognized and cofactors needed for the activity.
- Restriction enzymes derive their name from the genus, species and strains of bacteria from where they are originated, followed by a number that refers to discovery orders. For example; EcoR I and EcoR V are both from Escherichia coli, strain R; I and V are the order in which they were discovered.
How type II restriction enzymes works
- The Type II restriction enzymes recognize specific DNA sequences and cleave the DNA at fixed locations at or near the recognition sites. They act as dimers, each subunit recognizing the same 5 to 3′ nucleotide sequence in complementary DNA strands and hence are said to recognize palindromic sequences.
- Before the restriction enzyme cuts at its specific recognition site on a long DNA, it binds to a site amidst a very large number of non-cognate sites. This protein is then translocated from the initial site to a specific site, which occurs by “jumping or sliding”.
- At this recognition site in presence of Mg2+, enzyme undergoes a conformational change, which kinks the helix and cleaves the DNA producing ‘blunt’ or ‘sticky’ ends.
- For e.g. Hae III produced by Haemophilus aegypticus cleaves straight across the double helix when it encounters the following sequence to produce blunt ends.

- However, many restriction enzymes cut in an offset fashion to give sticky ends, which have protruding 5′ or 3′ ends with unpaired bases depending upon the restriction enzyme, e.g. EcoR I recognizes the following sequence and cleaves each backbone between G and A base residue giving 5′ protruding ends.

- Pst I recognizes the following sequence, cleaves between A and G residue to give 3′ protruding ends.
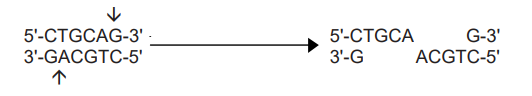
- The resulting sticky ends can base pair with any DNA molecule that has the complementary sticky ends to give a recombinant DNA molecule.
- When multiple enzymes recognize a single recognition sequence is known as isochizomers. For example Sma I and Xma I. These can cleave the DNA at same or different position within the recognition sequence.
- The frequency of cleavage varies on the possibility of the existence of recognition sequences. Therefore, an enzyme with longer recognition sequences cut less frequently and consequently produces large fragments than do enzymes with shorter recognition sequences.
Factors affecting Restriction Enzyme Activity
There are different factors that are influencing the enzyme activity such as;
- Temperature: Most of the digest is performed at 37°C. 37°C. However, there are a few exceptions e.g., digestion with Sma I is carried out at lower temperatures (~25°C), while with Taq I at higher temperature i.e., 65°C.
- Buffer Systems: Tris-HCl buffer is used as a buffering agent in incubation mixtures, which is temperature-dependent. Most restriction enzymes show activity at the pH range 7.0-8.0.
- Ionic Conditions: For the activity of restriction endonucleases Mg2+ ion is required. The requirement of ions (Na+/K+) varies based on enzymes.
- Methylation of DNA: Methylation of specific adenine or cytidine residues within the recognition sequence of the restriction enzyme affects the digestion of DNA.
Aim
- To perform restriction digestion of λ DNA with EcoR I and Hind III enzymes.
- To analyze the restriction pattern by agarose gel electrophoresis.
Restriction Digest Principle
Restriction Digestion involves fragmenting DNA molecules into smaller pieces with special enzymes called Restriction Endonucleases commonly known as Restriction Enzymes (RE). Because of this property restriction enzymes are also known as molecular scissors.
The restriction enzymes are named from the cellular strain from which they are isolated. Restriction enzymes recognize specific sequences in the double-stranded DNA molecule (for example GATATC) and then cut the DNA to produce fragments, called restriction fragments. The target site or sequence which the restriction enzyme recognizes is generally from 4 to 6 base pairs, arranged in a palindromic sequence.
Once it is located, the enzyme will attach to the DNA molecule and cut each strand of the double helix. The restriction enzyme will continue to do this along the full length of the DNA molecule which will then break into fragments. The size of these fragments is measured in base pairs or kilobase pairs (1000 bases).
Material Required for Restriction Digest
- 2X Assay buffer
- λ/Mlu I digest
- 2.5X Gel loading buffer
- Lambda DNA (substrate)
- Restriction Enzymes: EcoR I, Hind III
- Control λ DNA
- Agarose
- 6X Staining dye
- 50X TAE
- 1.5 ml vials
- Equipment: Dry bath, Gel rocker (optional).
- Glassware: Beakers, Conical flask, Measuring cylinder, Staining tray.
- Distilled water.
- Other: Crushed ice/Genei Cooler, Tips, Micropipette.
Restriction digest protocol
Preparation of agarose gel
- Prepare running buffer: In a clean two-liter container, add the entire contents of the TAE buffer (50X) and add two liters of ultra-pure water to make a 1X TAE buffer solution. Stir until thoroughly mixed.
- Prepare agarose: In a clean, glass 1000ml container add the entire contents of the agarose pack and add 500ml of the 1X TAE buffer from step 1.
- Heat the solution in a microwave on full power, using 10 second bursts, or use a boiling water bath. Check to see if all the agarose has dissolved. Continue until agarose has dissolved.
DO NOT BOIL. The agarose gets very hot, very quickly and can cause severe burns. Wear heat-protective gloves throughout the melting and pouring procedure.
- Once the agarose has cooled to the point it can be held comfortably in your hand, add the entire contents of the LabSafe™ Nucleic Acid Stain to the agarose and swirl to mix.
- Pour the agarose into the gel casting mould as per the manufacturer’s instructions. You will need 25 wells that each hold 30µl for each group, use an appropriate size comb.
- Once the gels have set, remove the comb, transfer to the running apparatus and cover with the running buffer until ready to use.
Restriction Digest
- Place the vials containing restriction enzyme (EcoR I and Hind III) on ice.
- Thaw the vials containing substrate (Lambda DNA) and assay buffer.
- Prepare 2 different reaction mixtures using the following constituents.
Reaction 1 (EcoR I digestion)
| λ DNA | 20 µl |
| 2X Assay Buffer | 25 µl |
| EcoR I | 3 µl |
Reaction 2 (Hind III digestion)
| λ DNA | 20 µl |
| 2X Assay Buffer | 25 µl |
| Hind III | 3 µl |
- Incubate the vial at 37°C for 1 hour.
- Meanwhile, prepare a 1% agarose gel for electrophoresis.
- After an hour add 5 μl of gel loading buffer to vials.
- Load the digested samples, 10 μl of Control DNA and 10 μl of marker, note down the order of loading.
- Electrophorese the samples at 50-100 V for 1-2 hours.
- Stain the agarose gel with 1X staining dye.
- Destain to visualize the DNA bands.
Observation and Result
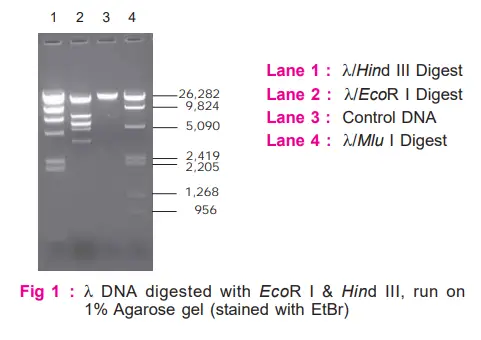
Observe the band patterns obtained on digestion with EcoR I and Hind III. Compare these with molecular weight marker (λ/Mlu I).
Interpretation
- Restriction patterns obtained on digestion with EcoR I and Hind III are markedly different, demonstrating the fact that each restriction enzyme recognizes and cuts only a particular base sequence unique to it.
- By comparing the migration distances with that of the marker, one can also determine the approximate sizes of DNA fragments.
Why is restriction digest important?
- Used for construction of recombinant DNA molecules
- Used to Mapping the locations of restriction sites in DNA
- Used in Southern Blot Hybridization
- Used in Construction of DNA Libraries
References
- https://en.wikipedia.org/wiki/Restriction_digest
- http://www.ispybio.com/search/protocols/RestrictionDigestion.pdf
- https://himedialabs.com/TD/HTBM015.pdf
- https://www.gbiosciences.com/image/pdfs/protocol/BE-307_protocol.pdf
- https://www.addgene.org/protocols/restriction-digest/
- https://www.sigmaaldrich.com/technical-documents/protocols/biology/restriction-enzyme-digest.html