What is Repetitive sequence-based PCR (rep-PCR)?
- The Repetitive sequence-based PCR (rep-PCR) was described as a Genomic Fingerprinting method, and it is mainly depended on amplification of DNA regions which are located between interspersed repetitive DNA elements in microbial genomes (REP, ERIC, BOX, GTG5 etc).
- In this technique, primers are designed that anneal to highly repetitive elements, and from these positions outward DNA fragments are amplified, so fingerprints are generated that appear as banding patterns.
- By the use of rep-PCR, Differentiation of bacterial isolates at species, subspecies and also Strain level can be obtained, and the produced patterns are often compared like “bar code” type which resemble UPC codes.
- In the definition, rep-PCR can be explained as a Polymerase Chain Reaction (PCR) based molecular typing tool, in which the distribution and orientation of conserved repetitive sequences are exploited for the production of complex genomic fingerprints.
- A rep-PCR genomic fingerprint is obtained when amplified fragments are separated on agarose gels or other electrophoresis system, and these fingerprints may then be analyzed either by human eye or by computer assisted approaches.
- In a more simple sense, rep-PCR is defined as a DNA fingerprinting methodology that utilize naturally occurring repetitive elements scattered throughout genomes, which are used as primer binding sites, and the variable distance between them is converted into unique electrophoretic profiles.
- For the understanding of rep-PCR, it should be noted that reproducible fingerprints are not only obtained from purified DNA but also from whole cells, colonies, crude lysates, and directly from plant lesions or nodules, which make the method widely applicable and also very flexible.
- The Definition is not limited only to bacteria, because rep-PCR has also been demonstrated to be useful in the study of fungi and other microorganisms, so its scope is extended beyond single domain.
- With the availability of improved versions like FERP (Fluorophore-enhanced repetitive PCR), the method has been further defined as adaptable to automation, multiplexing, and high-throughput analysis, which makes rep-PCR both simple and advanced at same time.
- Thus, rep-PCR is defined not just as one single PCR protocol but as a family of related protocols (REP-PCR, BOX-PCR, ERIC-PCR, GTG5-PCR) that all rely on interspersed repeats, and all together they are grouped under the general terminology rep-PCR genomic fingerprinting.
Working Principle of rep-PCR
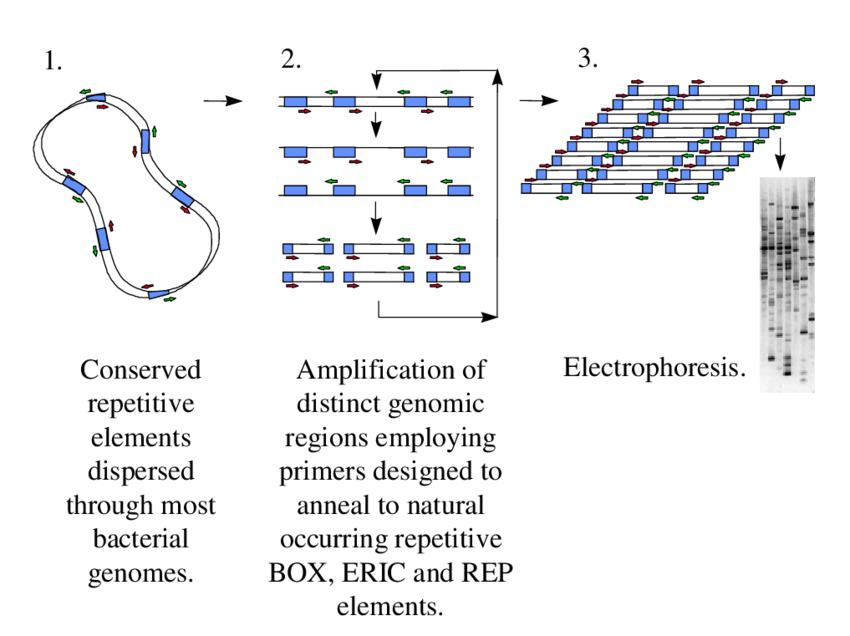
The rep-PCR working principle is based on the occurrence of conserved repetitive DNA elements (REP, ERIC, BOX, GTG5 etc.), which are dispersed at multiple sites within prokaryotic genomes, and these elements serve as priming sites for amplification.
By the use of specifically designed primers, annealing is allowed to repetitive elements that are found in both orientations, and then the DNA synthesis is primed outward, which results in amplification of the interspersed sequences located between them.
The amplified fragments are obtained in different sizes and lengths because the repetitive elements are unevenly distributed and variable in distance, so the resulting DNA patterns are highly complex.
During the PCR process, denaturation, annealing, and elongation steps are performed repeatedly, and each cycle is used to generate more amplified DNA regions flanked by these repetitive motifs.
The REP primer set, ERIC primers, BOX A1R, and also small polytrinucleotide primers like (GTG)5 are employed, but each primer system generates characteristic fingerprints, although differences in complexity and robustness are observed.
The produced amplicons are resolved by agarose gel electrophoresis, silver-stained polyacrylamide gels, or capillary electrophoresis, and the fragments are visualized as a banding profile that represent the genomic fingerprint of the isolate.
The resulting profiles are compared, and the differentiation of bacteria is obtained at species, subspecies or strain level, because the unique arrangement of repetitive elements in each genome is reflected in the banding pattern.
In rep-PCR, fingerprints are reproducibly generated not only from purified DNA, but also from crude cell lysates, colonies, and even from infected plant tissues, which illustrate the wide applicability of the method.
The working principle also includes optional modifications such as the use of fluorophore-labeled primers in FERP (Fluorophore-enhanced rep-PCR), where multiplex amplification and precise fragment sizing are made possible by laser-assisted detection.
A rep-PCR genomic fingerprint therefore works by converting the genomic organization of repetitive sequences into electrophoretic profiles, which resemble bar codes and can be studied by eye, or analyzed by software-assisted pattern comparison.1
Sample Requirement for rep-PCR
- The DNA purity must be high, and OD 260/280 ratio around 1.8 < OD < 2.0 is required, with no visible degradation or contamination.
- A minimum concentration of DNA is needed; often ≥ 10 ng/µl is acceptable for mixed microbial genomic DNA.
- Total amount of template DNA should be enough; more than about 1 µg of mixed microbial genomic DNA is often requested.
- For samples of Gram-positive bacteria, extra biomass is required, because their thick cell wall makes lysis / extraction harder; for community samples more than ~10 ml of microbial suspension (OD₆₀₀ > 1.5) or a dense microbial pellet is asked.
- The DNA must be intact (high molecular weight), and not smeared on gel, because degraded DNA gives poor band patterns or loss of resolution. (Inferred from purity / “no degradation” requirement)
- The concentration may need adjustment; for some protocols isolated DNA is normalized e.g. to ~50 ng/µl, so that amplification is reliable.
- The sample buffer / storage conditions should avoid inhibitors (e.g. salts, phenol, proteins) which might interfere with PCR; contaminants are to be minimized. (Inferred from requirement of “no contamination”)8
Protocol of rep-PCR
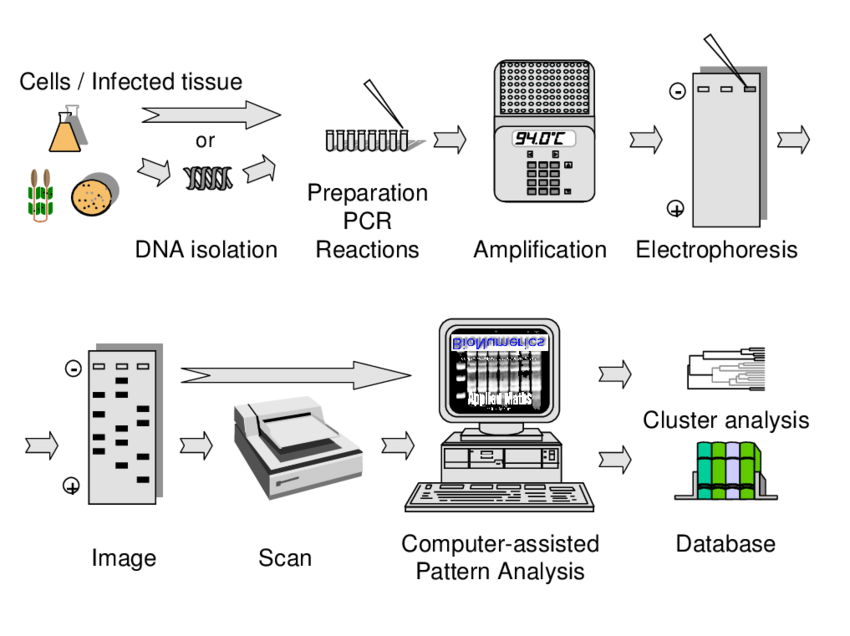
- The bacterial isolates are collected, and in many cases the growth is done overnight, so that sufficient biomass is obtained, which in turn ensure enough DNA yield for downstream.
- From the cultured cells, the genomic DNA was extracted, usually by enzymatic / chemical / commercial kit methods, and purity of DNA is checked, although sometimes the extraction give variable quality.
- A quantification of DNA is performed, by spectrophotometer or fluorometer, and the concentration is then adjusted, mostly in range of ng quantities, which is considered enough for PCR amplification.
- The primers are selected, and usually REP primers (REP-1R / REP-2I), ERIC primers (ERIC1R, ERIC2), or BOX A1R primer are being used, although sometimes species-specific primers are preferred by researchers.
- The PCR reaction mixture is prepared, containing buffer (Tris-HCl, KCl), MgCl₂, dNTPs, primers, thermostable DNA polymerase (like Taq), DNA template, and sterile water, which is finally adjusted to proper volume.
- In thermocycler, the program was set, and it generally include:
- Initial Denaturation at 94-95 °C (for 3-5 min)
- 30 to 35 cycles of denaturation at 94-95 °C (30-60 s), annealing (temperature around 40-55 °C depending primers) for 1-2 min, and extension at 72 °C for 1-2 min.
- Final extension at 72 °C (for 5-10 min), which is always included at the end.
- The control reactions are also included, like negative control (without DNA template), and molecular weight marker, which help in identification of band sizes.
- After the amplification, PCR products are separated by agarose gel electrophoresis, using concentration 1.5-2.5 % agarose in TAE/TBE buffer, and gels are run at voltages which sometimes vary depending on lab practice.
- The gel is stained, mostly with Ethidium Bromide or other safer dye, and after that, the visualization is done under UV or blue light transilluminator, so that banding patterns are made visible.
- The fingerprints are recorded, mostly by gel documentation system or by photographs, and these patterns are then compared among isolates.
- In analysis step, similarity coefficients are calculated, and dendrograms may be constructed, but sometimes only visual comparison of bands is used, which is less precise but still informative.
- The reproducibility of results is generally checked, by repeating the same rep-PCR on identical samples in different runs, and this ensures that patterns are stable and not just random amplification.23
Uses of rep-PCR
- The differentiation of microbial strains is being done by rep-PCR, and it is usually applied when the isolates of the same species are compared but they showing different banding patterns, which are often considered unique.
- For epidemiological investigations, rep-PCR has been widely applied, because the origin and spreading routes of infection could be identified, and sometimes the outbreak source is traced, even when other molecular tools were not used.
- In environmental samples, like soil / water / sewage sludge, the diversity of microorganisms has been evaluated by using the repetitive sequence based PCR method, although sometimes reproducibility is questioned, but still it is useful.
- The genetic relatedness of isolates has been assessed, and the dendrograms are constructed, from fingerprints, which, in turn, provide information about evolutionary distances between them, although the precision not always perfect.
- In the case of clinical outbreaks or food-borne contaminations, rep-PCR profiles are compared, so that clonality or non-clonality is confirmed, and by this, the infection dynamics can be more or less clarified.
- The population structure has been examined by researchers, where rep-PCR was performed for groups of bacterial species, and the intra-species variability has been highlighted, even though sometimes the conclusions are tentative.
- Industrial microbiology processes, like fermentation or starter culture production, are monitored, because rep-PCR is being applied for quality checking, ensuring that the strains are the same ones and not replaced by contaminants.
- For source tracking, contamination of environment by pathogenic bacteria has been investigated, and their origin (animal / human / environmental source) is determined, when fingerprints from different samples are compared side-by-side.
- Taxonomic clarification has been assisted, because rep-PCR was often used when subspecies classification was needed, especially when sequencing approaches were not affordable or too slow for practical laboratory settings.
- The monitoring of genetic stability has been carried out, as repeated rep-PCR testing over time may reveal rearrangements, insertions or deletions in repetitive sequences, and such variations are useful indicators of culture drift, although not always highly precise.
Advantages of rep-PCR
- The rep-PCR technique is considered highly rapid, since fingerprints of microbial isolates can be generated in only few hours, even though sometimes optimization is required before results appear clear.
- With relatively low cost of reagents and consumables, the method has been widely used, because expensive sequencing technologies are not always affordable, particularly in routine diagnostic or teaching laboratories.
- From very small quantity of template DNA, a large number of amplified products can be obtained, and therefore the method is seen as sensitive enough for different kinds of microbial samples.
- Intra-species differentiation has been successfully performed, since even closely related isolates are producing distinct banding patterns, which is a advantage when high resolution typing is demanded.
- The reproducibility of fingerprints has been reported in many studies, although slight variations might occur, but overall the patterns are considered reliable when same protocol is consistently applied.
- The technique is adaptable, and different primers (REP / ERIC / BOX or custom designed) can be selected according to organism of interest, so flexibility is given to researchers for multiple purposes.
- In comparison to other DNA based methods, rep-PCR require less technical expertise, and it can be done in ordinary molecular biology lab, without need of specialized or sophisticated equipment.
- A large number of isolates could be analyzed simultaneously, which save time and reduce work load, because many PCR reactions and gels can be run in parallel without major problems.
- The method has been applied for epidemiological tracking, strain identification, biodiversity assessment and also quality control, showing that one single approach can be used across diverse areas.
- The interpretation of results is relatively simple, as banding patterns are directly visualized on agarose gel, and comparison can be made visually or by computer software, giving both qualitative and quantitative options.
- The technique has been demonstrated to detect genetic variation, such as insertions, deletions or rearrangements within repetitive DNA sequences, which makes it useful for studies of genetic stability.
- Because standardized protocols have been already described in literature, the method can be reproduced and applied across different laboratories, even though some differences in gel conditions or annealing temperature may still be observed.4
Limitations of rep-PCR
- The reproducibility of banding patterns is often problematic, because small differences in PCR conditions / agarose gel electrophoresis / DNA quality can lead to shifts or missing bands, which reduce comparability.
- Between laboratories, standardization is difficult, since variation in primer sets, annealing temperatures, gel % (agarose concentration), buffer systems, run times produce inconsistent fingerprints, which makes inter-study comparisons unreliable.
- For organisms with low number of repetitive DNA elements (or evolutionary constrained repeats), discrimination is reduced, because few loci are available to produce distinct amplicon patterns, which limits the resolution.
- The presence of repetitive DNA itself causes amplification artifacts or failures, because mis‐priming, primer dimer formation, or slipped strand mispairing may occur, which results in spurious / missing or merged bands.
- With some rep-PCR primer sets, specificity is low for certain taxa, especially non-Gram negative or less studied species, because primers designed for one group may not perform well in others, which yields poor or ambiguous fingerprinting.
- High sensitivity to DNA template quality is required, since degraded, impure template or variation in DNA concentration affects band intensity / pattern, which may cause band loss or misleading differences.
- The method is not always as discriminatory as “gold-standard” typing methods (e.g. PFGE, whole genome sequencing), for distinguishing subtypes or outbreak strains, because subtle differences may be overlooked.
- Band scoring / interpretation is often subjective, when visual comparison is used, because small shifts in migration or faint bands are hard to judge uniformly, which introduces human error.
- Mixed populations or heterogeneity within sample may be masked, because if several colonies are pooled (or sample is composite), rare variants or minor genotypes are not detected, unless many individual colonies are tested.
- Some commercial kits / primer-sets may be discontinued or proprietary, making long-term continuity or reproducibility of studies difficult, because availability changes / protocol updates may occur.567
- Text Highlighting: Select any text in the post content to highlight it
- Text Annotation: Select text and add comments with annotations
- Comment Management: Edit or delete your own comments
- Highlight Management: Remove your own highlights
How to use: Simply select any text in the post content above, and you'll see annotation options. Login here or create an account to get started.