What is Pulsed-field Gel Electrophoresis (PFGE)?
Pulsed Field Gel Electrophoresis (PFGE) is a method used for separation of very large DNA fragments by applying an electric field that periodically changes direction / switches among different electrodes, so that DNA pieces of differing sizes are reoriented and separated. (The technique allows resolution of DNA up to several megabases, unlike normal gel electrophoresis which fails at > ~50 kb).
In PFGE, the electric field is pulsed (switched) so that large molecules that reorient slowly lag behind, and smaller ones reorient faster, so over time separation is achieved.
PFGE has been regarded as a “gold standard” for bacterial typing / genetic fingerprinting in epidemiology, outbreak studies, infection control etc because high discriminatory power is offered compared to many older methods.
Because PFGE is reproducible and patterns can be compared across labs (if standardized), it has been vital in tracking sources of disease, linking clinical isolates to environmental / food isolates, and in microbial population studies.
The first successful alternating-field method for large DNA separation was published in 1984 by David C. Schwartz and Charles Cantor, who applied this idea to separate chromosome-size DNAs.
After that, variations were developed (for example CHEF, OFAGE, TAFE etc) to refine performance, run time, direction switching schemes etc.
Principle of Pulsed Field Gel Electrophoresis (PFGE)
Large DNA molecules are loaded in agarose gel (or immobilized in agarose plugs) to avoid shearing, then electric field that periodically changes direction is applied so that DNA fragments reorient and migrate differently.
Because very big DNA fragments (> ~30-50 kb) under constant field move almost together (i.e. size-independent mobility), that regime is broken by pulsing, as different lengths respond to field switches at different speeds / with different lag.
When the field is switched, each DNA fragment must relax / reorient its conformation before it can move in the new direction; small fragments do it faster, large ones more slowly, so separation emerges over many pulses.
Pulses are applied between spatially distinct sets of electrodes (often at angles e.g. 60°) so that net migration is toward an anode although direction shifts, thus a zigzag or reorientation path is forced.
Over course of run (which may last many hours or days), fragments are separated based on ability to reorient under switch frequency / pulse times / field strengths.
The method depends on tuning of pulse time, field strength, angle and switching scheme to optimize resolution for given size range.
In short: alternating electrical fields force differential reorientation dynamics, and so large DNA pieces migrate more slowly than smaller ones in the pulsed regime, yielding size-based separation.
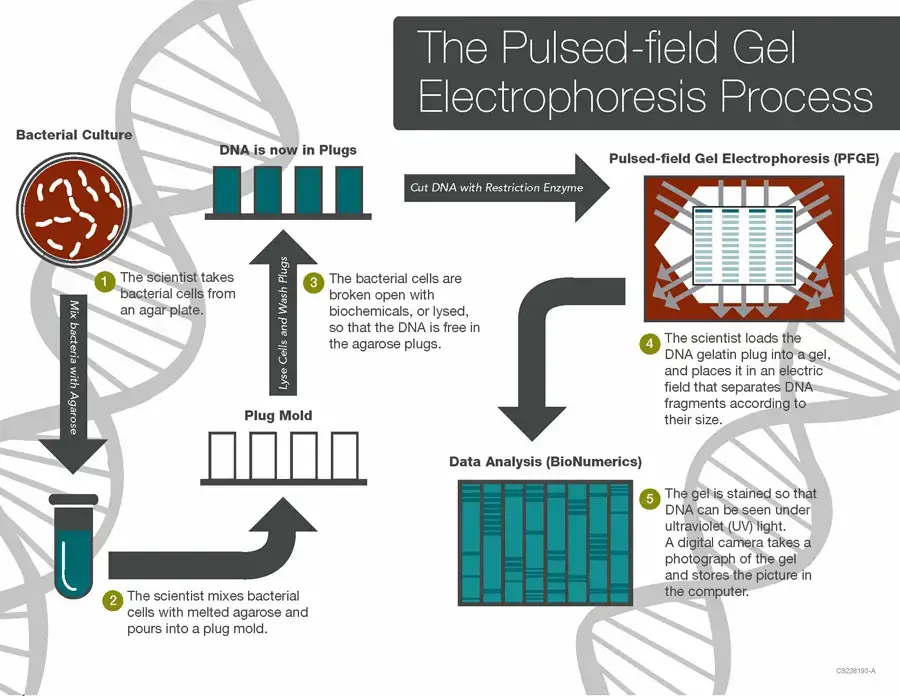
Procedure of Pulsed Field Gel Electrophoresis (PFGE)
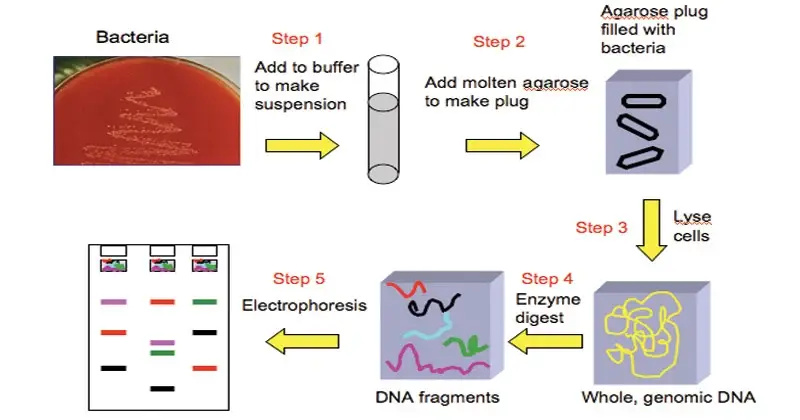
The major steps involved in Pulsed-field gel electrophoresis are;
- Cells are grown as pure culture (overnight etc) and harvested, to get enough biomass.
- Cells are suspended in buffer and mixed with molten low-melting agarose, then poured into plug mold so that cells become embedded in agarose plugs.
- The agarose is allowed to solidify (set) so that DNA inside is immobilized and less subject to mechanical shearing.
- Lysis of cells is done while inside the plugs, with enzymes / detergents / proteinase K etc, so that cellular membranes / proteins are removed and intact chromosomal DNA is freed inside agarose.
- The plugs are washed multiple times with buffer (e.g. TE) to remove lysis reagents and residual contaminants.
- A slice (e.g. ~2 mm thick) is cut from each plug, using sterile scalpel / blade, for restriction digestion.
- The slice is incubated with a rare-cutting restriction enzyme (with buffer and co-factors) so that the embedded DNA is digested in situ to yield large fragments.
- Prepare agarose gel (often low percent agarose) and wells; insert plug slices into wells (and seal wells sometimes with agarose).
- PFGE is performed: electric field is applied and periodically switched among certain directions (e.g. 0°, ±60°) so that DNA fragments reorient and migrate differentially.
- Electrophoresis is run over long period (many hours to even >24 h) under controlled temperature (buffer circulated, cooling, etc) to avoid overheating.
- After run, gel is stained (e.g. ethidium bromide or safer dye) to visualize DNA bands.
- Gel is de-stained / washed, then photographed or scanned (e.g. gel doc) under UV light.
- Banding patterns are analyzed (manually or by software like BioNumerics) and compared across samples for relatedness / typing.
Applications of Pulsed Field Gel Electrophoresis (PFGE)
- PFGE is mostly applied for molecular typing of bacteria, it helps in identifying and comparing different strains in epidemiological studies.
- It is used for tracking source of infection during outbreak investigations (like foodborne diseases by Salmonella, Listeria monocytogenes etc.).
- The method is used by public health labs like CDC PulseNet network for monitoring and tracing foodborne pathogens, their distribution and spread.
- PFGE is applied to study genetic diversity within bacterial population and between isolates from different origins / environments.
- The technique is used for genome mapping to analyze large chromosomal DNA fragments which can’t be resolved by normal gel electrophoresis.
- Chromosomal rearrangements / mutations in microorganisms are detected by this method, providing information about genomic stability.
- It is widely used for strain differentiation and genotyping among closely related microbial isolates, which is crucial for taxonomy and diagnostics purpose.
- PFGE has been used for determining genome size of prokaryotic and eukaryotic organisms, since it separates DNA fragments above 50 kb accurately.
- It is employed for characterization of antibiotic resistant bacteria, showing genetic variation or transfer of resistance genes among isolates.
- In research, PFGE used for clonal relationship analysis to know whether isolates from various patients or locations belong to same lineage or not.
- It has application for detecting plasmid or bacteriophage integration events within bacterial chromosomes.
- PFGE results are often used for comparative genomics studies and evolutionary relationship interpretation among species etc.
- Some applications also involve validation of recombinant strains, checking for unwanted chromosomal rearrangements after genetic manipulation.
- In environmental microbiology, PFGE is used to analyze microbial communities from soil, water, and food surfaces for contamination studies.
- Overall PFGE serves as a gold standard technique for microbial genotyping, even though new molecular methods like WGS are emerging, PFGE still used due to its reliability and cost-effectiveness.
Advantages of Pulsed Field Gel Electrophoresis (PFGE)
- A major advantage is that very large DNA fragments (up to several Mb) can be separated accurately by PFGE.
- It is considered as highly reproducible method, which make it suitable for comparison between different laboratories.
- PFGE gives high resolution separation pattern, so even small genetic differences among closely related strains are detected.
- The method has been widely used for epidemiological typing, since the results are stable and can distinguish between strains of same species.
- Genome mapping of bacteria, yeast, and fungi is made possible by PFGE due to its ability to resolve large chromosomal fragments.
- It can be used for identifying genetic rearrangements / mutations, which can’t be easily seen by standard electrophoresis techniques.
- PFGE allows analysis of intact chromosomal DNA without the need of prior fragmentation or cloning.
- The technique is relatively simple once standardized, and can be performed in most molecular labs with basic equipment.
- Since it uses restriction enzymes, banding patterns are specific and reliable for strain identification.
- PFGE results are highly interpretable visually, and databases can be maintained for pattern comparison (like in PulseNet).
- It is useful for determining genome size of organisms with high accuracy.
- The method has wide applicability for both prokaryotic and eukaryotic cells (bacteria / fungi / protozoa).
- Compared with other typing methods, PFGE is cost-effective and time saving for large-scale outbreak analysis.
- It provides permanent DNA fingerprint, that can be stored digitally for future references.
- Despite new sequencing technologies, PFGE still remain as gold standard for microbial subtyping due to its precision, reliability and universality.
Limitations of Pulsed Field Gel Electrophoresis (PFGE)
- PFGE procedure is time-consuming, usually taking 2–4 days for completion including DNA preparation and electrophoresis run.
- It requires specialized equipment and technical skill, so not all labs can perform it easily or consistently.
- The DNA preparation step must be done very carefully because chromosomal DNA is fragile and easily sheared during handling.
- Standardization problems often occur between laboratories, small changes in protocol may cause large variations in band patterns.
- PFGE can’t resolve small DNA fragments (<50 kb) efficiently, hence it not suitable for short sequence analysis.
- The method is labor-intensive and require high quality reagents / restriction enzymes that sometimes are costly.
- Sometimes band interpretation becomes difficult due to co-migration of fragments or unclear bands.
- Data analysis is manual and subjective, since minor differences in migration can lead to wrong interpretation.
- PFGE patterns are not easily comparable with sequence-based data, so integration with modern genomic databases is limited.
- The technique is less discriminatory for some species where genetic variability is low, giving same pattern for unrelated strains.
- DNA degradation or incomplete digestion often affect result reproducibility, leading to false clustering of isolates.
- It is not ideal for real-time outbreak tracking, since result generation take longer compared to PCR or sequencing-based typing.
- PFGE cannot detect single nucleotide changes (SNPs) or small mutations, so it has lower resolution compared with WGS.
- Large electric field setups may cause gel distortion or overheating, especially during long runs (above 24–48 hrs).
- Overall, PFGE though powerful, is being gradually replaced by faster, high-resolution, sequence-based approaches due to these practical limitations.
FAQ
What is Pulsed-field Gel Electrophoresis (PFGE)?
PFGE is a technique used to separate and analyze large DNA fragments, typically ranging from a few kilobases to several megabases, based on their size using an electric field.
What is the purpose of PFGE?
PFGE is primarily used for molecular fingerprinting and genotyping of bacteria, allowing researchers to study genetic relatedness, identify outbreaks, and trace the source of infections.
How does PFGE differ from traditional gel electrophoresis?
Unlike traditional gel electrophoresis, PFGE uses alternating electric fields at regular intervals to separate large DNA fragments, providing enhanced resolution and the ability to analyze fragments that are too large for conventional methods.
What are the key steps involved in PFGE?
The major steps in PFGE include preparing the agarose gel, loading the DNA sample, running the electrophoresis with pulsed electric fields, and staining the gel to visualize the separated DNA fragments.
What are the advantages of PFGE?
PFGE allows for the separation of large DNA fragments, provides high resolution, is versatile for different bacterial species, and offers stable and reproducible results.
What are the limitations of PFGE?
Some limitations of PFGE include being time-consuming, requiring trained technicians, limited discrimination between unrelated isolates, slight result variations between technicians, and the inability to type certain strains.
Can PFGE be used for genotyping other organisms besides bacteria?
Yes, PFGE can be used for genotyping other organisms with large genomes, such as fungi, protozoa, and even mammals. It is a versatile technique for analyzing the structure and origin of genomes.
Is PFGE a widely accepted technique in epidemiological studies?
Yes, PFGE is considered a gold standard in epidemiological studies of pathogenic organisms due to its high discriminatory power and ability to link isolates from outbreaks and infections.
What factors can be optimized to enhance PFGE results?
Factors that can be optimized for better PFGE results include voltage, pulse angle, switch time, and temperature, depending on the size of the DNA fragments being analyzed.
Can PFGE be combined with other molecular techniques?
Yes, PFGE can be combined with other techniques such as DNA sequencing, PCR, and hybridization methods to further analyze and characterize DNA fragments of interest.
- Sharma-Kuinkel BK, Rude TH, Fowler VG Jr. Pulse Field Gel Electrophoresis. Methods Mol Biol. 2016;1373:117-30. doi: 10.1007/7651_2014_191. PMID: 25682374; PMCID: PMC4582012.
- Parizad EG, Parizad EG, Valizadeh A. The Application of Pulsed Field Gel Electrophoresis in Clinical Studies. J Clin Diagn Res. 2016 Jan;10(1):DE01-4. doi: 10.7860/JCDR/2016/15718.7043. Epub 2016 Jan 1. PMID: 26894068; PMCID: PMC4740595.
- https://thesciencenotes.com/pulsed-field-gel-electrophoresis-pfge/
- https://www.bio-rad.com/en-in/applications-technologies/pulsed-field-gel-electrophoresis?ID=LUSORPDFX
- https://www.sciencedirect.com/topics/medicine-and-dentistry/pulsed-field-gel-electrophoresis
- https://www.applied-maths.com/applications/pulsed-field-gel-electrophoresis-pfge-typing
- https://microbeonline.com/pulsed-field-gel-electrophoresis-pfge/
- https://international.neb.com/tools-and-resources/video-library/what-is-pulsed-field-gel-electrophoresis
- https://experiments.springernature.com/articles/10.1385/0-89603-498-4:33