What is Peripheral Protein?
- Peripheral protein, or peripheral membrane protein, is a collection of physiologically active amino acid-based molecules that interact with the lipid bilayer surface of cell membranes.
- Peripheral proteins, unlike integral membrane proteins, do not enter the hydrophobic region within the cell membrane.
- Instead, peripheral proteins include particular amino acid sequences that enable them to bind to the phosphate heads of lipid molecules or to integral proteins.
- Peripheral proteins are able to function on the surface of the cell membrane because they can bind to the membrane without being locked to it.
- Numerous mechanisms allow for the activation or deactivation of peripheral proteins.
- Numerous peripheral proteins are also involved in numerous intricate metabolic processes.
- They may be involved in the movement of substances within or beyond a cell, the activation of other proteins and enzymes, or cell-to-cell contacts.
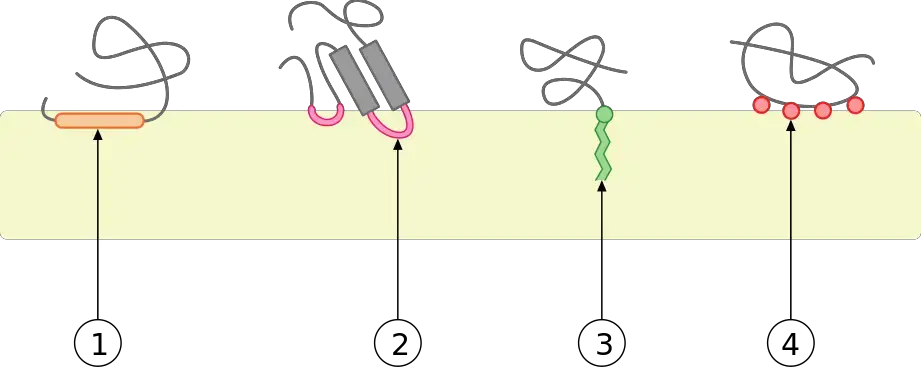
Structure of Peripheral Proteins
Several peripheral proteins are marked in the picture below. A peripheral protein doesn’t have a clear structure, but it does have a few important features that make it what it is.
- All proteins on the outside of the cell are connected to the cell membrane. The sequences of amino acids in these proteins are unique in that they attract the proteins to the membrane, where they tend to gather.
- This puts them in the right place to do what they are supposed to do. In the picture, you can see that the orange proteins on the outside are attached to either the phosphoglyceride lipid molecules that make up the lipid bilayer or to the proteins in the middle.
- Without these parts of amino acids, a protein would not stick to the membrane. It would be spread out evenly in the cytoplasm and not be a protein on the edges.
- Second, amino acids that don’t like water are not found in the outer parts of proteins. This, along with the fact that some amino acid groups are polar, keeps the proteins on the outside of the cell membrane.
- This is because phosphoglycerides are both good and bad. This means that the blue “head” is polar and has a high affinity for water.
- The yellow “tails” in the middle of the membrane are not able to hold water.
- Proteins on the outside of the membrane often have a lot of hydrophilic amino acids on their surface so they don’t get sucked into the membrane.
- Integral proteins have amino acids that are not affected by water in the middle and amino acids that are affected by water on the outside. This keeps them locked up inside the membrane.
Example of Peripheral Proteins
Alternative Oxidase
- Alternative oxidases (AOXs) are terminal quinol oxidases that are attached to membranes and are found in the respiratory electron chains of many species from different kingdoms.
- Since there is no human equivalent for AOXs, they have been studied in depth as drug targets for dangerous fungi and the human parasite Trypanosoma brucei (T. brucei).
- African trypanosomiasis, also called “sleeping sickness,” is caused by parasites called T. brucei. If it isn’t treated, it will kill you.
- AOXs are very important to the way these pathogens use energy, so they have become a new therapeutic target for treating diseases caused by these pathogens.
- AOXs speed up the process of turning oxygen into water, even in the presence of cyanide, without the need to move protons across the inner mitochondrial membrane.
- By doing this, they act as a part of the respiratory electron transport chain that doesn’t save energy.
- Iron is important for AOXs to work well. So, to be active, AOXs must interact with oxygen, a membrane, quinols, and iron.
- Crystal structures of AOXs showed that these enzymes are homodimers, with six -helices in each monomer. Four of these -helices come together to make a 4-helix bundle, which holds two iron atoms together.
- The active site of the enzyme is this four-helix bundle with a diiron in the middle.
Cytochrome c
- Cytochrome c (cyt c) comes in two forms: one that is attached to the cell membrane and one that is soluble.
- Cyt c is a small, highly basic hemoprotein that is found in the space between the membranes of mitochondria (the cytoplasmic membrane of bacteria). In human mitochondria, it has two biological functions.
- The main job of cyt c is to help move electrons from cyt c reductase, which is part of the bc1 complex, to IMP cytochrome c oxidase.
- The first step of this reaction is for the soluble cyt c to move toward the membrane-bound COX. This is made easier by the long-range electrostatic interaction between cytochrome c and the membrane.
- The release of cyt c from the mitochondria into the cytosol starts the apoptotic pathway. This is the second biological function of cyt c.
- When cyt c binds to the inner membrane of the mitochondria, it not only speeds up the transfer of electrons in the respiratory chain, but it also keeps the cell from dying in a process called apoptosis.
- It has been thought for a long time that the permeability transition pore is how mitochondrial cyt c gets out.
- The dimeric F1Fo ATP synthase, which is part of the respiratory chain that cyt c is a part of, has recently been linked to the permeability transition pore. This is a new discovery that may help explain how cyt c gets out of mitochondria.
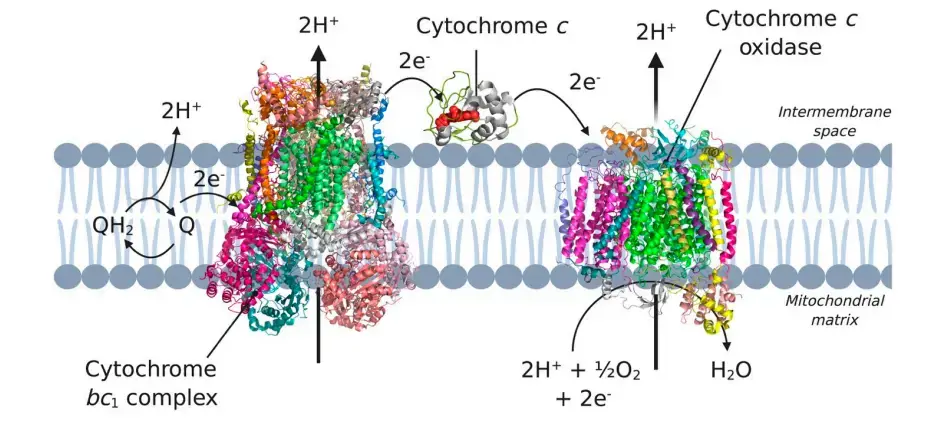
- But cyt c is also an interesting target in, for example, cancer treatment because the release of cyt c from mitochondria into the cytosol starts the apoptosis pathway.
- Targeted delivery of cyt c to cancer cells is now thought to be a possible way to cause apoptosis and kill the cancer cells.
- As was already said, cyt c needs to be drawn to the membrane so that it can do its main job in the body. Cardiolipin, one of the lipids that make up the mitochondrial membrane, helps this attraction happen (CL).
- CL has a unique structure because it has a small head group but four acyl chains. This phospholipid is a big part of the inner membrane of the mitochondria and is needed for proteins to stay in place, stay stable, and work normally.
- Deprotonated or partially protonated cardiolipin forms a complex with cyt c through electrostatic interactions, but possibly also through hydrogen bonding and hydrophobic interactions.
- This unique lipid interacts with cyt c mostly at two acidic phospholipid binding sites on the surface called the A- and C-sites.
- It is thought that deprotonated CL molecules interact with cyt c through electrostatic interaction at the A-site, and that proteins bind to protonated phospholipids through hydrogen bonds at the C-site.
- The interactions between CL and cyt c not only determine where cyt c binds to the membrane, but they also change the shape of the protein when it binds to lipids and cause structural changes in the lipid bilayer that are caused by the protein.
- Along with the unique parts of CL’s structure, a hydrophobic crack was found in cyt c’s tertiary structure. This led to the idea that a so-called “extended lipid conformation” could be made when a molecule binds to a membrane.
- In this shape, the acyl chains of the lipids point in the opposite direction of the head group, making a right angle of 180°.
- One of the CL acyl chains stays in the lipid bilayer, and the other goes out. Then, this growing acyl chain fits into a space in cyt c that doesn’t like water.
- It is thought that this anchorage causes conformational changes in cyt c, such as loosening of the tertiary structure, changes in the haem environment, and reversible unfolding.
- When cyt c binds to the membrane, it changes how the lipids in the inner mitochondrial membrane are organised. In particular, cyt c binding causes changes in the way CL is distributed, which leads to the formation of microdomains that are rich in CL.
- Multiple studies have shown that putting cyt c in a CL domain speeds up the rate at which the protein transfers electrons. CL domains may also make the apoptotic signal stronger.
Type-II NADH Dehydrogenase
- Type-II NADH dehydrogenases (NDH-2s) are NADH:quinone oxidoreductases that are very important in the respiratory metabolism of bacteria and in the mitochondria of fungi, plants, and protists.
- They speed up the two-step transfer of an electron from an aqueous NADH substrate, through their cofactor FAD, to a membrane-bound quinone substrate, most often ubiquinone or menaquinone.
- Unlike the NADH dehydrogenases found in humans (called type-I NADH dehydrogenases or respiratory complex I), type-II NADH dehydrogenases do not pump protons across the membrane. Instead, they only add to the membrane’s electrical potential ().
- They don’t exist in humans, so they make great drug targets against many different pathogens, from Staphylococcus aureus to Mycobacterium tuberculosis.
- People have said that these enzymes are “peripheral membrane proteins” that are found in the inner mitochondrial membrane that faces the cytosol or in the bacterial cell membranes.
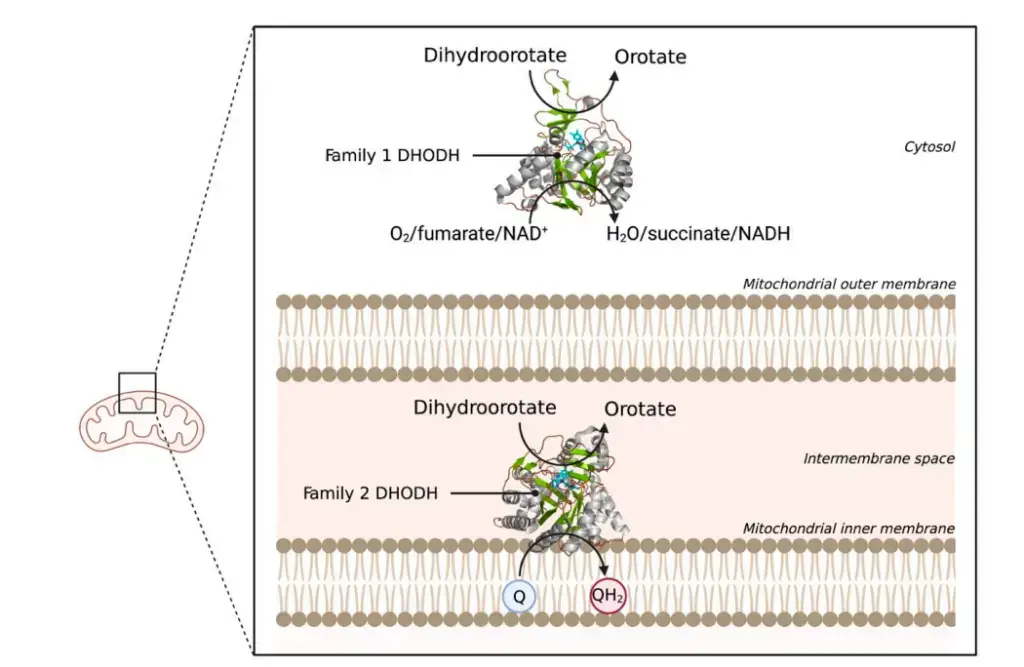
Dihydroorotate Dehydrogenase
- The enzyme dihydroorotate dehydrogenase (DHODH) is the last example of an electrostatically linked peripheral membrane protein. It has been shown to be a possible target in the fight against cancer.
- DHODH is needed for de novo pyrimidine biosynthesis, which is needed to make DNA and RNA, which is needed to make more cells.
- The de novo pyrimidine biosynthesis route isn’t used much in healthy cells, but it is the main source of pyrimidines in cells that divide quickly, like tumour cells.
- Because of this, stopping DHODH could possibly lead to the loss of pyrimidines in tumour cells.
- But it’s still not clear if the lack of pyrimidines could cause the response of the p53 tumour suppressor and, in the end, cause p53-mediated cell death.
- There are two groups of DHODHs: family 1 and family 2.
- Family 1 DHODHs are found in the cytosol and accept electrons from oxygen, fumarate, or NAD+.
- Family 2 DHODHs are bound to membranes and accept electrons from ubiquinones.
- DHODH speeds up two redox reactions: the change from dihydroorotate to orotate and then the reduction of ubiquinone.
- The oxidation of dihydroorotate is a step-by-step process that uses residues that are very stable.
- A catalytic base residue is used to remove the proton from the C5 position of DHO. A hydride is then moved to a flavin mononucleotide (FMN).
- In the second reaction, a two-electron transfer needs to happen between FMN and a ubiquinone from the electron transport chain.
- The electrons go to the membrane-bound acceptor ubuquinone-10 via the cofactor FMN (Q10). So, to stop DHODH from working, the coenzyme Q10 binding site in the amphipathic domain can be filled in a specific way.
- The N-terminal region, which has a mitochondrial signal sequence, a putative transmembrane helix, and two amphipathic helices, is what connects the protein to the membrane.
- The membrane-binding domain, which helps move two electrons from the soluble substrate dihydroorotate, is in the amphipathic region. This area is amphipathic, which means it can move in both directions. It interacts with lipids to position the protein and bind it to the membrane.
Functions of Peripheral Proteins
1. Provide Support
- One of the main jobs of peripheral proteins is to guide and maintain both the cytoskeleton inside the cell and the parts of the extracellular matrix that are outside the cell.
- Both of these things are made up of many organelles, filaments, and tubules. These small structures can give stiffness or tension, but they have to be attached to something.
- This point of attachment to the cell membrane can be made by proteins on the outside of the cell. Both the cytoskeleton and the extracellular matrix are used in many ways by cells. Most of the time, they are used to control how the cell looks and how big it is.
- The cytoskeleton also helps move around products of metabolism and can be stopped or started by proteins on the outside of the cell.
- For example, a new package of proteins made in the Golgi apparatus could use the cytoskeleton to move through the cytosol.
- When the package gets to the cell membrane to be thrown out, certain proteins on the outside of the cell recognise it and start the process of throwing it out.
2. Help in Communication
- In addition to supporting the structure of the body, the extracellular matrix is also a large network that many cells use to share information.
- Bacteria, for example, use a series of reactions that start in the filaments of their extracellular matrix to turn on proteins on the outside of their cells.
- These proteins then send the message to proteins that are part of the cell, which takes the message inside the cell.
- Here, it is sent to another protein on the outside, and then a response is started. This is a good way for a tiny organism or cell to learn a lot about its surroundings.
- This is how cells that are growing together to make a multicellular organism respond and know when to stop growing.
- Peripheral proteins and many other proteins and chemical signals set off chains of reactions that can make the DNA or other organelles do something.
- So, a cell can grow more, respond to a threat, or even make its own toxins based on its microenvironment and the signals it gets.
- Also, many proteins on the outside of the membrane can attach and detach from it based on things like pH and temperature. This lets a cell adapt to different environments and control things like how cells talk to each other and how they respond to hormones.
3. Act as an Enzyme
- There are many proteins on the outside of cell membranes that do something to a certain substrate.
- This could be done to break it up or put it together with another molecule.
- Peripheral proteins with simple enzyme functions are often peripheral proteins because the molecules they make are needed inside or near the cell membrane.
- For example, several enzymes that control how the cell membrane is made and broken down are peripheral proteins.
4. Hel in Molecule Transfer
- Many peripheral proteins are also involved in transferring small molecules or electrons.
- These proteins, due to their affinity to the cell membrane, allow the reactions to stay in a tight space, and be highly coordinated.
- Many of the proteins found within the electron transport chain are peripheral proteins.
- These proteins transfer electrons from integral proteins they are attached to, and can pass the electrons to other proteins and molecules.
- Effectively, this stores the energy from the breakdown of the products of glycolysis into easily accessible molecules, or ATP.
- Other molecules, which are hydrophobic, can bind to peripheral proteins and be passed through various methods across or through the membrane.
References
- Cavaliere, Matteo & Sedwards, Sean. (2007). Membrane Systems with Peripheral Proteins: Transport and Evolution. Electronic Notes in Theoretical Computer Science. 171. 37–53. 10.1016/j.entcs.2007.05.006.
- Boes, D.M.; GodoyHernandez, A.; McMillan, D.G.G. Peripheral Membrane Proteins: Promising Therapeutic Targets across Domains of Life. Membranes 2021, 11, 346. https://doi.org/10.3390/ membranes11050346
- Sixma, J. J., Schiphorst, M. E., Verhoeckx, C., & Jockusch, B. M. (1982). Peripheral and integral proteins of human blood platelet membranes α-actinin is not identical to glycoprotein III. Biochimica et Biophysica Acta (BBA) – Protein Structure and Molecular Enzymology, 704(2), 333–344. doi:10.1016/0167-4838(82)90163-7
- https://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.1069.3225&rep=rep1&type=pdf
- https://www.ck12.org/book/ck-12-biology-advanced-concepts/section/3.12/
- https://biologywise.com/difference-between-peripheral-integral-membrane-proteins
- https://www.creative-biolabs.com/blog/index.php/membrane-protein-overview/
- https://en.wikipedia.org/wiki/Peripheral_membrane_protein
- https://www.biologyonline.com/dictionary/peripheral-membrane-protein
- https://www.sciencedirect.com/topics/biochemistry-genetics-and-molecular-biology/peripheral-membrane-protein
- https://ib.bioninja.com.au/standard-level/topic-1-cell-biology/13-membrane-structure/membrane-proteins.html
- https://biologydictionary.net/peripheral-proteins/