Northern blotting is the process used for detecting specific RNA molecules from a mixture of total RNA, and it is mainly applied for studying gene expression. It is the method where RNA is first isolated from cells or tissues, and this step is very sensitive because the RNA easily gets degraded if RNase is present.
The extracted RNA is then separated according to size by using denaturing agarose gel electrophoresis. In this step formaldehyde is added in the gel, preventing the formation of any secondary structure so the migration of RNA occur based only on size. After the separation, the RNA present in the gel is transferred onto a nylon membrane by capillary action or vacuum blotting and the membrane holds the RNA firmly. The RNA is then fixed on the membrane by UV cross-linking or by heating.
The membrane is kept in pre-hybridization solution which helps in reducing non-specific binding. In the next step, a labeled probe which is single-stranded DNA or RNA is allowed to hybridize with the complementary RNA sequence that is present on the membrane. The probe can be radioactive or non-radioactive depending on the detection system.
After hybridization the membrane is washed to remove unwanted binding and the hybrid signal is visualized. It is the technique that helps in determining the amount of a particular RNA, the size of the transcript and the pattern of expression.
Northern blotting is important because it can directly show the transcript size, detect splice variants, and also indicate the RNA integrity by showing the 28S and 18S ribosomal RNA bands.
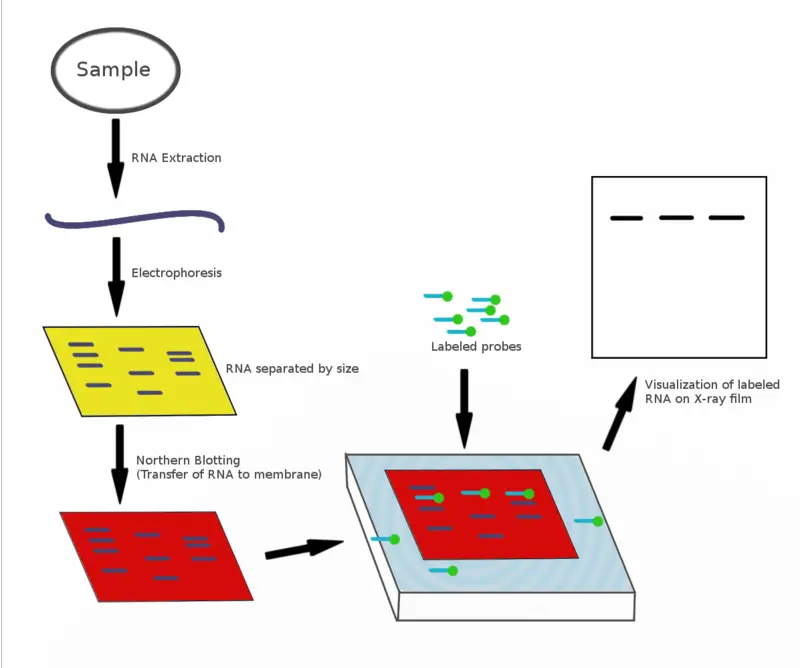
Principle of Northern Blot
The principle of Northern blot is based on the separation of RNA molecules according to their size and the specific hybridization between the RNA and a complementary probe. It is the process where the RNA sample is first run on a denaturing agarose gel.
These denaturing conditions are necessary because RNA is single-stranded and it easily forms secondary structures, so the denaturant helps in keeping the RNA in a linear form during electrophoresis. The separated RNA fragments are then transferred from the gel onto a nylon membrane because RNA binds strongly to nylon, and nitrocellulose is not preferred for this purpose. After transfer, the RNA is immobilized on the membrane by UV-cross linking or by heating so that it remains fixed during the next steps.
The detection is based on nucleic acid hybridization. A labeled single-stranded probe is added which is complementary to the target RNA sequence present on the membrane. This is the step that gives specificity because only the probe with complementary bases binds to the desired RNA molecule.
After washing to remove any non-specific binding, the probe–RNA hybrid is visualized. It is the principle that allows identification of the RNA segment, its size, and relative expression because the migration on gel is size dependent and the probe binding confirms the exact sequence present in the sample.
Material Required for Northern Blotting
Equipment
- Agarose gel cast (used for preparing the gel for RNA separation).
- Power supply for providing voltage during electrophoresis.
- Microwave for melting agarose.
- Centrifuge which is used for separation of components in sample preparation.
- Heating block for maintaining constant temperature in different steps.
- UV crosslinker as it is used to fix RNA on membrane.
- Hybridization oven for providing uniform condition during hybridization.
- Hybridization vessels which hold membrane and probe.
- Vials used for preparing and storing solutions.
- Forceps for handling membranes without contamination.
- Pipettes for accurate liquid transfer.
- Glass tubes used during reactions and hybridization.
Materials
- Agarose gel which is the medium for RNA separation.
- Sodium citrate used as buffering agent.
- EDTA (Ethylenediaminetetraacetic acid disodium salt dehydrate) for chelating metal ions.
- NaOH (Sodium hydroxide) for denaturation of RNA.
- HCl (Hydrochloric acid) used in denaturation steps.
- Formaldehyde which is used as denaturing agent.
- Glycerol added for increasing density of sample.
- Ethidium bromide used to visualize RNA under UV.
- Bromophenol blue which acts as tracking dye.
- RNA ladder used as size marker.
- MgCl₂ used in buffer preparation.
- NaCl used commonly in buffers.
- PVP (Polyvinylpyrrolidone) for preventing non-specific binding.
- BSA (Bovine serum albumin) used for stabilizing enzymes.
- SDS (Sodium dodecyl sulfate) used as detergent.
- NaH₂PO₄ used as buffering agent.
- Tris-HCl (Tris(hydroxymethyl)aminomethane hydrochloride) for buffer preparation.
- Triton-X used as non-ionic detergent.
- DTT (Dithiothreitol) which acts as reducing agent.
- Taq buffer used for maintaining reaction conditions.
- Taq polymerase enzyme used for amplification of nucleic acid.
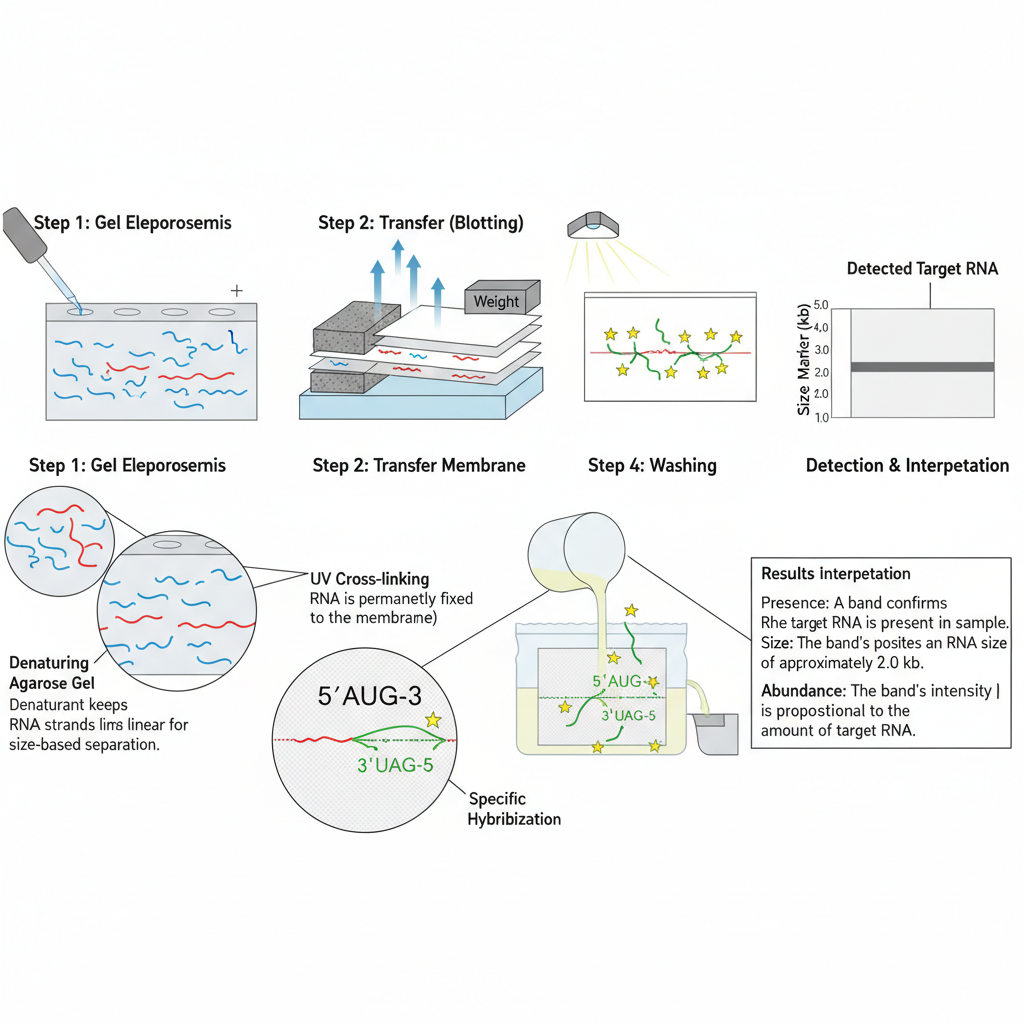
Northern Blotting Steps/Procedure
Northern blotting is the method used for detection of specific RNA molecules. It is the process where RNA is separated on denaturing gel, transferred to membrane and hybridized with a labeled probe. These steps are carried out in RNase-free condition because RNA is easily degraded.
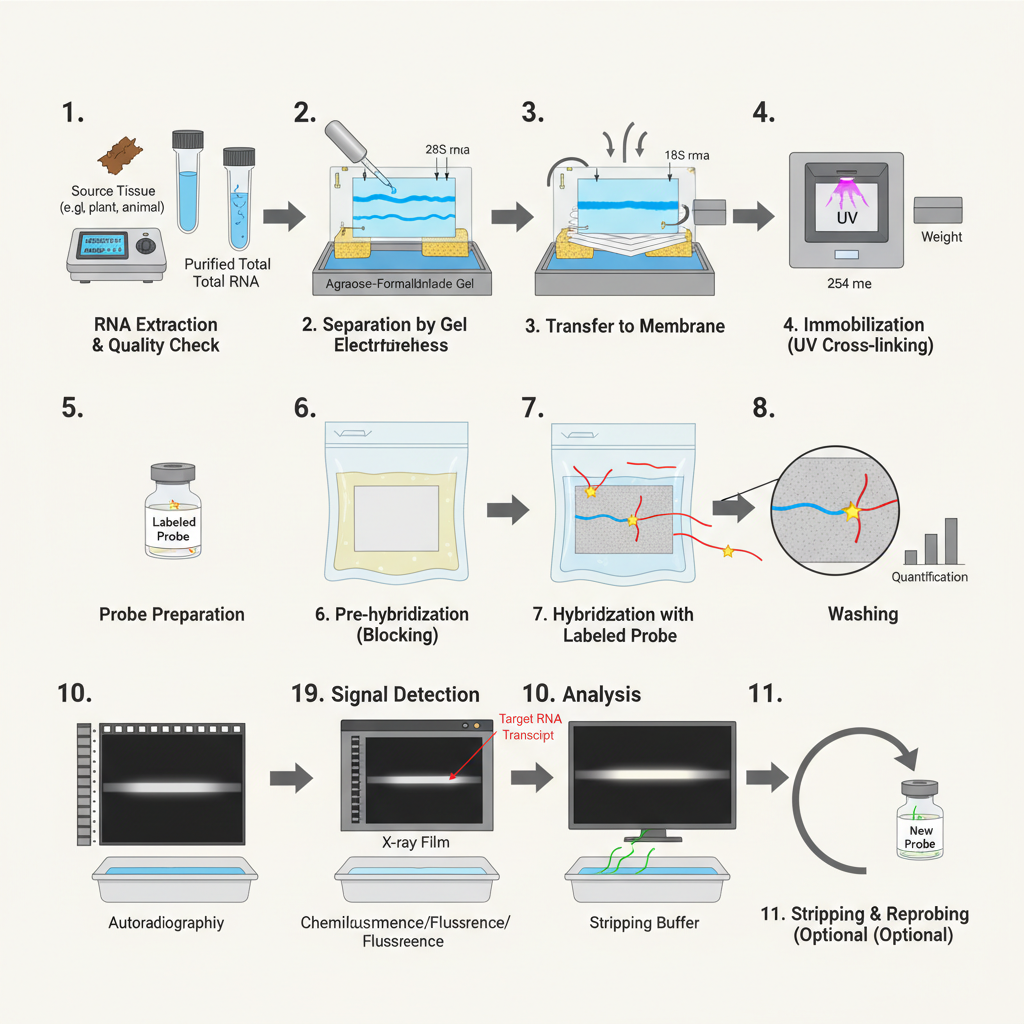
1. RNA Extraction and Quality Checking
It is the initial step where the total RNA is isolated from tissue or cells. The extraction is usually done by phenol-chloroform method or silica column procedures. The isolated RNA is treated with DNase so that any contaminating DNA is removed.
It is essential to maintain RNase-free conditions because RNA gets degraded very fast. The quality of RNA is checked by spectrophotometer (A260/A280) or by running a small amount on gel. These are used to confirm that the RNA is not degraded.
Sometimes mRNA is isolated from total RNA using oligo-dT beads when only gene expression study is required.
Steps
- Collection of sample – Cells or tissue are collected and kept in cold condition to prevent RNA degradation. It is the stage where RNase contamination is avoided.
- Lysis of cells – The sample is mixed with lysis buffer containing phenol, chloroform or guanidinium salts. It is the process which breaks open the cells and denatures proteins including RNases.
- Separation of phases – After lysis, the mixture is centrifuged. The aqueous phase containing RNA is separated from the organic phase. This is referred to as phase separation.
- Precipitation of RNA – The aqueous phase is transferred and RNA is precipitated using isopropanol or ethanol. It forms a visible pellet after centrifugation.
- Washing of RNA pellet – The pellet is washed with 70% ethanol so that salts and impurities are removed. The pellet is then air-dried for a short time.
- Dissolving the RNA – The dried pellet is dissolved in RNase-free water or TE buffer. It is essential that all solutions are RNase-free.
- DNase treatment – The extracted RNA is treated with DNase enzyme. This is the step removing any contaminating DNA so that only RNA remains.
- Checking the purity – RNA purity is checked by spectrophotometer. The A260/A280 ratio between 1.8–2.1 indicates good quality RNA.
- Checking the integrity – A small amount of RNA is run on agarose gel. Two clear bands of 28S and 18S rRNA indicate that the RNA is intact.
- Storage – The purified RNA is stored at −80°C or in liquid nitrogen so that degradation does not occur.
2. Separation of RNA on Denaturing Gel
The RNA samples are separated based on size. It is done on agarose-formaldehyde gel as formaldehyde keeps the RNA in denatured form.
The denaturing gel is prepared by adding formaldehyde to agarose solution and poured into the cast. When the gel sets, the wells are formed by removing the comb.
The gel is equilibrated with running buffer for around 30 minutes. The RNA sample (usually 15 µg) is mixed with RNA loading buffer and heated at 65°C for about 12–15 minutes. RNA markers are also added.
In this step, the samples are loaded into the gel and run at around 125V. It is the process where 28S and 18S bands become visible after staining which confirms the integrity of RNA.
Steps
- Preparation of denaturing gel – Agarose is dissolved in buffer and formaldehyde is added. It is the process that keeps RNA in denatured form by preventing secondary structure formation.
- Casting of gel – The warm gel solution is poured into the gel tray. A comb is inserted to form wells, and the gel is allowed to set properly.
- Equilibration of gel – After solidification, the comb is removed and the gel is placed in running buffer for about 30 minutes. This step allows the gel to equilibrate with the buffer.
- Preparation of RNA samples – RNA sample (usually 15 µg) is mixed with RNA loading dye. The mixture is heated at 65°C for 12–15 minutes so that the RNA is fully denatured before loading.
- Addition of RNA markers – A separate tube containing RNA marker is prepared with loading dye. These markers are used to determine the size of transcripts.
- Loading of samples – The equilibrated gel is placed in the electrophoresis tank. RNA samples and markers are carefully loaded into the wells using RNase-free tips.
- Running of gel – Electrophoresis is carried out at around 125V. It is the step where RNA fragments move according to size in the presence of formaldehyde.
- Visualization check – After the run, the gel can be stained using ethidium bromide or other stains. The appearance of sharp 28S and 18S rRNA bands indicates good RNA integrity.
3. Transfer of RNA to Membrane (Blotting)
The separated RNA is transferred from the gel to a nylon membrane. Nylon membrane is used because it has high binding capacity for nucleic acids.
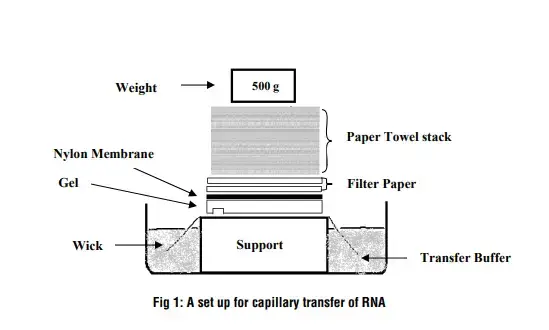
For capillary transfer, a sponge soaked in SSC is taken and covered with wet filter papers. The gel is placed on the filter paper and any air bubbles are removed.
The membrane is wetted in distilled water and then kept over the gel carefully. Filter papers and a glass plate are placed above it. It is kept overnight so that RNA is transferred completely to the membrane.
Steps
- Preparation of membrane and papers – A nylon membrane is cut slightly larger than the gel. Filter papers of the same size are also prepared. The membrane is wetted in distilled water on an RNase-free dish for a few minutes.
- Setting up the transfer platform – A sponge is placed in a glass tray and soaked with SSC buffer. The sponge is kept half-submerged so that continuous upward flow of buffer occurs during transfer.
- Placement of filter papers – Two or more Whatman 3mm filter papers are placed on the sponge. These papers are soaked properly with SSC to maintain even contact.
- Positioning of the gel – The agarose gel containing separated RNA is removed from the tank and rinsed gently with water. The gel is then placed on the wet filter papers. Air bubbles are removed by rolling a glass pipette over the surface.
- Laying the nylon membrane – The pre-wetted nylon membrane is placed directly on top of the gel. It is essential that no air bubbles remain between the gel and the membrane because this interrupts transfer.
- Stacking additional filter papers – A few dry filter papers are placed over the membrane. These papers draw the buffer upward, pulling RNA from the gel onto the membrane.
- Applying weight for uniform contact – A clean glass plate is kept on top of the stack. A small weight may be added to ensure even pressure and proper capillary action throughout the set-up.
- Incubation period – The entire arrangement is allowed to stand undisturbed, usually overnight. It is the step where RNA molecules slowly migrate from the gel and bind to the nylon membrane.
- Completion of transfer – After transfer, the membrane is removed, briefly rinsed in SSC, and air-dried. The gel is discarded or kept for checking transfer efficiency.
4. Immobilization of RNA
After transfer, the membrane is dried and fixed so that the RNA is permanently attached. It is usually done by baking the membrane at 80°C for 2 hours. In some cases, the membrane is exposed to UV light which forms covalent bonding with RNA. This is referred to as cross-linking.
Steps
- Drying the membrane – After the RNA transfer is complete, the nylon membrane is removed from the transfer setup and rinsed briefly with SSC. It is then allowed to air-dry so that excess moisture is removed.
- Placing between filter papers – The dried membrane is positioned between two clean filter papers. This arrangement protects the membrane surface and keeps it flat during the fixing process.
- Thermal baking – The membrane is placed in a vacuum oven at 80°C for around 2 hours. It is the step where heat creates stable linkage between the RNA and membrane.
- Alternative UV cross-linking – In some procedures, instead of baking, the membrane is wrapped in UV-transparent film and exposed to 254 nm UV light. This exposure forms covalent bonds between the RNA and membrane.
- Cooling and storage – After baking or UV cross-linking, the membrane is cooled and stored in a dry, RNase-free container. It is the process ensuring that the RNA remains tightly fixed for hybridization.
5. Hybridization with Labeled Probe
In this step, a labeled DNA or RNA probe complementary to the target RNA is prepared. The probe is labeled either radioactively or by non-radioactive tags.
The membrane is pre-hybridized first with blocking solution to reduce non-specific binding. Then the hybridization solution containing the probe is added.
If the probe is double-stranded, it is denatured by heating at around 100°C for 10 minutes. The membrane is incubated in the hybridization oven, usually at 42°C or required temperature, until the probe binds to the target RNA.
Steps
- Preparation of the probe – A labeled DNA or RNA probe is prepared which is complementary to the target RNA. The probe is labeled with radioactive or non-radioactive tags, and the unincorporated nucleotides is removed. It is the step ensuring that only active probe is present for hybridization.
- Wetting of membrane – The membrane containing the fixed RNA is first wetted with SSC buffer. This helps the membrane to take up the hybridization solution evenly.
- Pre-hybridization – The membrane is kept in a hybridization tube with the RNA-side up. A pre-hybridization or blocking solution is added to the tube. It is the process which reduces any non-specific binding on the membrane surface.
- Incubation for blocking – The tube is placed in a hybridization oven and incubated at about 42°C for a few hours. This step allows proper blocking of the membrane before adding the probe.
- Denaturation of probe – If the probe is double-stranded DNA, it is denatured by heating at 100°C for around 10 minutes. This converts the probe into single strands so that it can bind to the target RNA.
- Addition of probe – The required amount of denatured probe is added into the hybridization tube. The solution is mixed gently so that the probe spreads uniformly over the membrane.
- Hybridization step – The tube is again incubated at 42°C or at the specific temperature required. It is the step where the probe anneals to the complementary RNA present on the membrane.
- Removal of solution – After the hybridization period, the solution is poured off. The membrane is then ready for washing to remove any loosely bound probe.
6. Washing of Membrane
Unbound or loosely bound probes are removed by washing the membrane using different wash solutions.
Low-stringency washes remove loosely attached probes whereas high-stringency washes remove non-specific hybridization. It is the step ensuring that only the correct probe-RNA hybrid remains.
Steps
- Initial rinse of membrane – After hybridization, the membrane is taken out of the tube and given a quick rinse with a basic wash solution. It is the step removing loosely attached probe from the membrane surface.
- Low-stringency wash – The membrane is placed in a container containing low-stringency wash buffer. This buffer removes probe molecules that are weakly bound. The membrane is incubated at a moderate temperature with gentle shaking.
- Replacement of wash buffer – The low-stringency solution is poured off and replaced with fresh buffer if required. This helps in removing the remaining nonspecific bindings.
- High-stringency wash – The membrane is transferred to a high-stringency wash buffer. It is the process where more stringent conditions (higher temperature or lower salt) are applied to remove probe molecules that are not perfectly complementary.
- Incubation in high-stringency conditions – The membrane is incubated for the required time so that only highly specific probe-target hybrids remain. This step ensures clear and accurate signal formation.
- Final rinse – The membrane is given a final rinse with SSC or the recommended buffer to remove traces of wash solution. It prepares the membrane for detection.
- Drying of membrane – The washed membrane is air-dried or blotted between filter papers. It is then ready for the detection step.
7. Detection
The membrane is then exposed to X-ray film (autoradiography) or phosphorimager screen depending on label type.
Dark bands appear on the film which shows the transcript. These bands are used to estimate the size and quantity of the RNA. The amount is usually normalized against ribosomal RNA bands or housekeeping gene signals.
Steps
- Preparation for detection – After washing, the membrane is placed on a clean surface or tray. It is ensured that the membrane is free from excess buffer because moisture can interfere with signal development.
- Selection of detection method – The method depends on the type of label used on the probe. Radioactive probes are detected by autoradiography, while non-radioactive probes are detected by chemiluminescent or fluorescent systems.
- Exposure to X-ray film (for radioactive probes) – The membrane is wrapped in plastic wrap and placed against an X-ray film inside a cassette. It is kept at low temperature to allow clear band formation. This is the process where radiation from the probe exposes the film.
- Using phosphor imaging screen (alternative method) – For phosphor screen detection, the membrane is kept in close contact with the screen. After exposure, the screen is scanned in a phosphor imager to obtain digital signals.
- Development of film – When autoradiography is used, the X-ray film is developed in a darkroom. The bands appear as dark regions where hybridization occurred.
- Observation of signal – The membrane or developed film is examined for the presence of bands. These bands represent the target RNA molecules hybridized with the labeled probe.
- Recording of results – The band intensity and size are recorded. This data is used for further analysis and comparison with RNA markers or internal controls.
8. Stripping and Reprobing (Optional)
The membrane can be stripped by removing the bound probe through specific solutions.
It is reused for hybridization with another probe so that more than one transcript can be studied from the same blot.
Steps
- Preparation of membrane for stripping – After detection, the membrane is placed in a clean container. Any remaining moisture or film residues is removed gently so that the surface is ready for the stripping solution.
- Addition of stripping solution – A specific stripping buffer is added to the container. It is the solution that removes the previously bound probe from the membrane without damaging the RNA fixed on it.
- Incubation in stripping conditions – The membrane is incubated at the recommended temperature, often warm conditions. This allows the probe to detach from the RNA strands on the membrane.
- Rinsing after stripping – The stripping buffer is poured off, and the membrane is rinsed several times with SSC or suitable wash solution. It removes leftover probe fragments and prepares the membrane for the next hybridization.
- Checking membrane readiness – The membrane is checked visually to ensure that no signal from the previous probe is present. It is important so that the next probe does not overlap with earlier signals.
- Pre-hybridization again – Before using a new probe, the membrane undergoes pre-hybridization to block non-specific sites. This step restores the membrane to the same condition as before the first hybridization.
- Reprobing with new labeled probe – A fresh labeled probe complementary to another target RNA is added. The same hybridization procedure is followed. It allows the study of multiple transcripts on a single membrane.
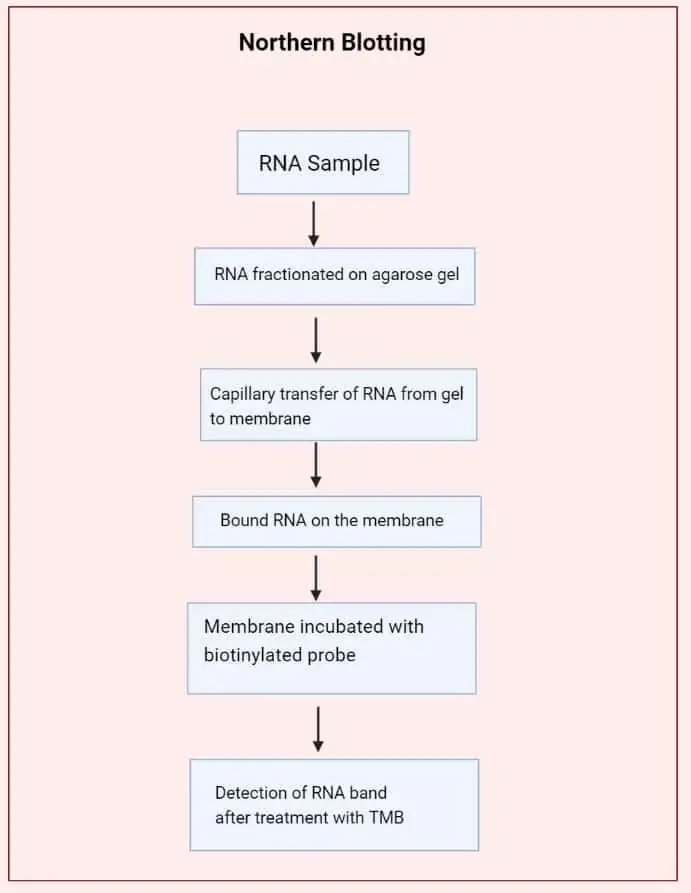
Result Interpretation of Northern Blot
The interpretation of Northern blot results is based on the observation of bands formed on the membrane after hybridization with the specific probe. It is the process where the presence, size, and amount of the RNA transcript is understood from these bands. The major points for interpretation are as follows–
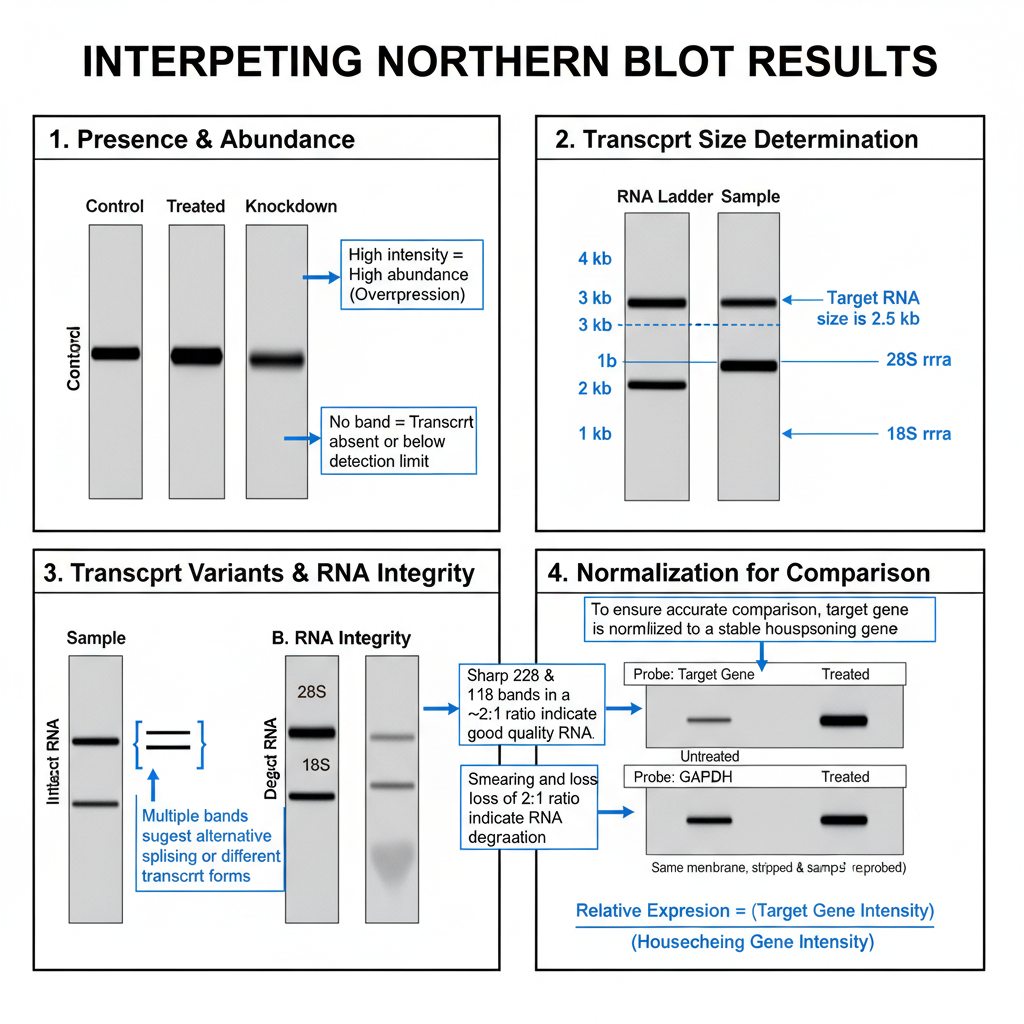
1. Presence of Transcript and Its Abundance
A clear band appearing at the expected position indicate the presence of the target RNA. If no band is observed, then it shows that the transcript is absent or present in very low amount. The intensity of the band is directly related to how much of that RNA is present in the sample.
When different samples are compared side-by-side, the difference in band intensity indicate overexpression or underexpression of that gene. It is observed in studies involving treated and untreated cells or different tissues. Quantification is usually done by densitometry where the signal intensity is measured.
2. Determination of Transcript Size
Since RNA is separated on the denaturing gel according to size, the position of the band reflects the molecular size of the transcript. It is compared with RNA ladder. In total RNA, the 28S and 18S rRNA bands appear as internal size indicators. The band lying near a particular marker shows its approximate length. This is one of the important advantages of Northern blot.
3. Detection of Transcript Variants and RNA Integrity
Sometimes more than one band hybridizes with the same probe. It is the indication that the gene has alternative splice forms or different polyadenylation products. It can also arise due to degradation intermediates.
The RNA quality must be checked initially. Intact RNA show sharp 28S and 18S rRNA bands with nearly 2:1 ratio. A smear below these bands or loss of this ratio indicate degradation. In such cases the result becomes unreliable.
4. Normalization for Comparison
The target band intensity is normalized with an internal control to obtain proper comparison between samples. It is usually normalized with housekeeping genes like GAPDH or β-actin, or with the 28S and 18S rRNA bands. The value obtained after dividing the target signal with the control signal is used to compare relative expression.
Some genes like EF-1α (Elongation factor-1α) show stable expression and can serve as good internal control in some plant experiments.
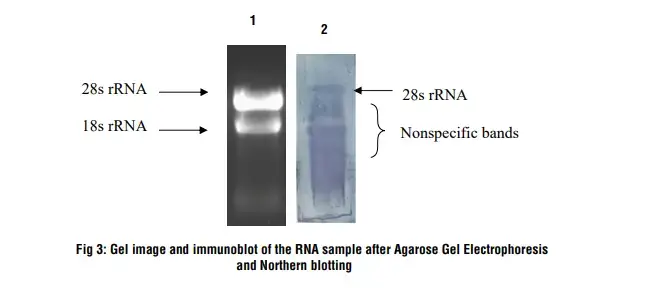
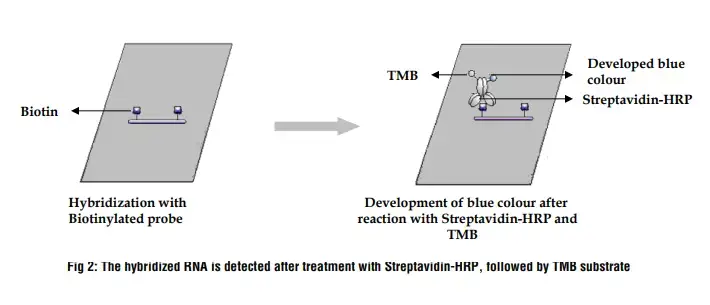
Lane 1: RNA sample on 1.2 % denaturing agarose gel
Lane 2: After Northern hybridization a blue band develops on the nylon membrane
Precautions
- All steps must be carried out in RNase-free condition because RNA is easily degraded.
- RNase-free tubes, tips, glassware and solutions are used and work surface is cleaned properly.
- Gloves must be worn and changed frequently since fingers can introduce RNase contamination.
- The extracted RNA should be checked on denaturing gel to confirm clear 28S and 18S rRNA bands with proper 2:1 ratio.
- Denaturing gel must contain formaldehyde or other denaturant to prevent formation of secondary structures.
- The transfer membrane must be handled carefully so that RNA is not lost before cross-linking.
- Proper UV cross-linking or heating at around 80°C is needed to fix RNA permanently on the membrane.
- Pre-hybridization is required to block non-specific binding sites on the membrane.
- Hybridization and washing steps must follow correct stringency so that only specific probe–RNA binding remains.
- Chemicals like formaldehyde and ethidium bromide must be handled in fume hood due to their toxicity.
- If radioactive probes are used, proper radiological safety rules, disposal and monitoring must be followed.
- All steps must be performed carefully to avoid background signal and loss of sensitivity in the final blot.
Northern blotting vs Southern blotting
| Northern Blotting | Southern Blotting | |
|---|---|---|
| Target Molecules | RNA molecules | DNA molecules |
| Purpose | Detection and analysis of RNA molecules | Detection and analysis of DNA molecules |
| Technique | Gel electrophoresis, membrane transfer, hybridization | Gel electrophoresis, membrane transfer, hybridization |
| Target Sequence | Complementary DNA (cDNA) or RNA probes | DNA probes |
| Applications | Gene expression analysis, RNA processing studies, non-coding RNA analysis | DNA analysis, gene mapping, detection of specific DNA sequences |
| Detection Method | Radioactive or non-radioactive labeling of probes | Radioactive or non-radioactive labeling of probes |
| Sensitivity | Generally lower sensitivity compared to newer RNA analysis methods such as qRT-PCR or RNA-seq | Can detect low-abundance DNA sequences, high sensitivity possible with appropriate probes and conditions |
| Sample Requirements | Requires a relatively large amount of RNA sample | Requires a relatively large amount of DNA sample |
| Limitations | Time-consuming, lower sensitivity, limited dynamic range for quantification | Time-consuming, potential for DNA degradation, limited dynamic range for quantification |
| Key Applications | Gene expression analysis, mRNA splicing analysis, non-coding RNA analysis | DNA mapping, detection of specific DNA sequences, analysis of DNA mutations or polymorphisms |
Application of northern blotting
- It is used to study gene expression by detecting the presence and amount of specific mRNA in different samples.
- It help in observing the expression pattern of a gene in different tissues, organs, developmental stages or during stress conditions.
- It is applied to compare overexpression or underexpression of genes in diseased cells like cancer cells.
- It is used to determine the exact size of the RNA transcript because RNA is separated according to size before hybridization.
- It help in detecting alternative splicing forms when more than one band is observed for the same probe.
- It is useful in studying degradation products and processing intermediates of RNA.
- It is used to check RNA integrity by observing clear 28S and 18S rRNA bands on the gel.
- It is applied to validate results obtained from RT-qPCR and RNA-Seq because it provide direct non-amplified evidence of transcript level.
- It help in confirming newly predicted splice variants or transcript variants by detecting their size difference on the blot.
Advantages of Northern Blotting
- It determines the exact size of the RNA transcript because separation occurs on denaturing gel before detection.
- It help in identifying alternative splice forms when more than one band hybridizes with the same probe.
- It can show transcript processing errors or missing regions by using probes from different parts of the gene.
- It provides a clear RNA integrity check through visible 28S and 18S rRNA bands with expected 2:1 ratio.
- It has high specificity and reduces the chance of false positive signals.
- It gives direct non-amplified measurement of transcript abundance, making it useful for validating RT-qPCR data.
- It allows the use of probes with partial homology, including cDNA or genomic DNA fragments.
- It is considered reliable and cost-effective for gene expression analysis in many laboratory conditions.
- It allows reprobing of the same membrane after stripping the old probe, increasing data obtained from a single sample.
- It accepts different types of probes like DNA probes, RNA probes and oligonucleotide probes, providing flexibility in hybridization.
Limitations of Northern Blotting
- It is a slow and labor-intensive technique because each step like electrophoresis, transfer and hybridization take long time.
- It is low-throughput and not suitable for analyzing many genes or many samples at the same time.
- It has low sensitivity and cannot detect transcripts present in very low amount.
- It requires high quantity of RNA sample, usually in microgram level.
- The quantification obtained from band intensity is less precise compared to RT-qPCR.
- The RNA used must be completely intact because even slight degradation produce smear instead of clear band.
- RNA is easily destroyed by RNases, so the risk of degradation is always present during the procedure.
- Formaldehyde, ethidium bromide, DEPC and UV exposure used in the process are harmful chemicals.
- Use of radioactive probes require strict handling, disposal and safety precautions.
- Detection of multiple transcripts needs stripping and reprobing, which takes more time and the membrane can only be reprobed limited times.
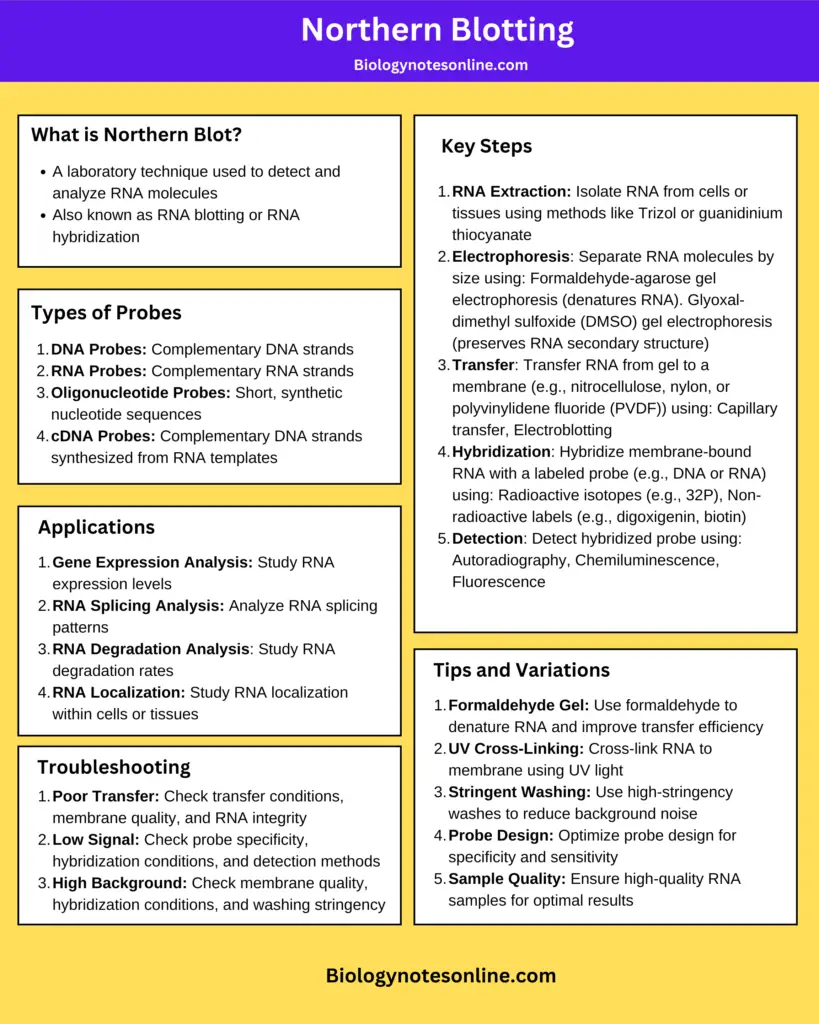
Northern Blotting Cheatsheet/Infographic
FAQ
What is Northern blotting?
Northern blotting is the technique used to detect specific RNA molecules. It is the process where RNA is separated on denaturing gel, transferred to a membrane and hybridized with a labeled probe.
What is the primary purpose of Northern blotting?
The primary purpose is to study gene expression by observing the presence and amount of specific RNA transcripts.
How does Northern blot differ from Southern blot?
Northern blot is used for RNA, while Southern blot is used for DNA. The separation in Northern blot is done on denaturing RNA gel, whereas Southern blot separates DNA fragments.
What are the steps involved in Northern blotting?
The steps include RNA extraction, separation of RNA on denaturing gel, transfer to membrane, immobilization, probe hybridization, washing and detection.
What kind of information can be obtained from Northern blotting?
It provide information about presence of transcript, its size, abundance, splice variants and RNA integrity.
How is the RNA sample separated in Northern blotting?
The RNA sample is separated on denaturing agarose gel containing formaldehyde which prevent formation of secondary structure.
What is the role of the probe in Northern blotting?
The probe is complementary to the target RNA sequence. It binds to the RNA on the membrane and helps in detecting the specific transcript.
How is the separated RNA transferred to a membrane in Northern blotting?
The RNA is transferred by capillary blotting or vacuum blotting where the RNA moves from gel to the nylon membrane.
What type of gel is used in Northern blotting?
A denaturing agarose gel containing formaldehyde is mostly used.
What is the solid support (membrane) typically used in Northern blotting?
Nylon membrane is commonly used because it has high affinity for nucleic acids and good mechanical strength.
How is the target RNA sequence visualized after hybridization in Northern blotting?
It is visualized by autoradiography if radioactive probe is used or by chemiluminescent or fluorescent detection when non-radioactive probes are used.
What are the advantages and disadvantages of Northern blotting?
Advantages include determination of transcript size, detection of splice variants, high specificity and direct non-amplified quantification.
Disadvantages include low sensitivity, requirement of high RNA amount, time-consuming steps and risk of RNA degradation.
How can I avoid background or non-specific bands in Northern blotting?
Proper pre-hybridization, correct hybridization temperature and stringent washing help in reducing non-specific binding.
How can I prevent RNA degradation during Northern blotting?
Using RNase-free reagents, wearing gloves, cleaning surfaces and handling all materials in RNase-free condition help in preventing degradation.
What are the applications of Northern blotting?
It is used to study gene expression, detect splice variants, analyze RNA processing, check RNA integrity and validate results from RT-qPCR or RNA-Seq.
- Boster Bio. (2024, February 15). A Guide to 10 Blotting Types.
- Chakravarty, P. R. (2025). An Overview of Northern blot. GoldBio.
- Dean, J. D., Goodwin, P. H., & Hsiang, T. (2002). Comparison of relative RT-PCR and northern blot analyses to measure expression of β-1,3-glucanase in Nicotiana benthamiana infected with Colletotrichum destructivum. Plant Molecular Biology Reporter, 20(4), 347–356.
- He, S. L. (2013). Northern blot. Methods in Enzymology, 530, 75–87.
- National Human Genome Research Institute. (2025, December 1). Northern Blot. Genome.gov.
Northern Blotting: A Comprehensive Analysis of Methodology, Applications, and Comparative Role in Modern Transcriptomics. (n.d.). - Sigma-Aldrich. (n.d.). Northern & Southern Blot Protocols.
- Tebubio. (2023, May 11). Southern – Northern – Western Blot : what is the difference?.
- Thermo Fisher Scientific. (n.d.). The Basics: Northern Analysis.
- Wikipedia contributors. (n.d.). Northern blot. In Wikipedia.
- https://himedialabs.com/TD/HTBM028.pdf
- https://www.thesciencenotes.com/northern-blotting-principle-procedure-and-applications/
- https://www.mybiosource.com/learn/testing-procedures/northern-blotting/