What is Protein Synthesis Regulation?
Protein synthesis regulation refers to the various mechanisms by which cells control the production of proteins. Protein synthesis is the process by which cells generate proteins, which are essential molecules for the structure, function, and regulation of various cellular processes.
The regulation of protein synthesis allows cells to adjust protein production in response to internal and external signals, ensuring that the cell produces the right proteins at the right time and in the right amounts. It is crucial for maintaining cellular homeostasis, responding to environmental changes, and coordinating cellular activities.
There are multiple levels of regulation involved in protein synthesis, including transcriptional, post-transcriptional, translational, and post-translational mechanisms. Here’s a brief overview of each level:
- Transcriptional regulation: This level of regulation controls the synthesis of messenger RNA (mRNA) molecules, which serve as templates for protein production. Transcription factors and regulatory proteins bind to specific DNA sequences and either enhance or repress the transcription of genes, thereby influencing the amount of mRNA produced.
- Post-transcriptional regulation: After transcription, mRNA molecules undergo various modifications and processing steps before they are ready for translation. This includes the addition of a 5′ cap and a poly(A) tail, as well as the removal of introns through splicing. Regulatory elements, such as RNA-binding proteins and non-coding RNAs, can interact with mRNA molecules and influence their stability, localization, and translational efficiency.
- Translational regulation: At the translational level, the process of protein synthesis is controlled. Various factors, including initiation factors, ribosomes, and regulatory proteins, can modulate the efficiency and rate of translation. For example, the binding of specific proteins or non-coding RNAs to the mRNA can promote or inhibit translation, ensuring selective protein synthesis.
- Post-translational regulation: Once proteins are synthesized, they can undergo further modifications that affect their stability, activity, localization, and interactions with other molecules. These modifications include phosphorylation, acetylation, glycosylation, and proteolytic cleavage, among others. Post-translational modifications can alter the function or fate of a protein and play crucial roles in cellular signaling and regulation.
Overall, protein synthesis regulation is a complex and highly coordinated process that allows cells to fine-tune protein production in response to changing conditions. It ensures the proper functioning of cells and contributes to the overall control of cellular processes.
Translation Steps in Eukaryotic Cells
Translation is the process by which the information carried by mRNA molecules is used to synthesize proteins. In eukaryotic cells, translation occurs in the cytoplasm and involves several steps. Here are the key steps involved in translation in eukaryotic cells:
- Initiation: The small ribosomal subunit binds to the mRNA molecule at the 5′ cap region. The ribosome scans the mRNA until it reaches the start codon (usually AUG) that signals the initiation of translation. Initiator tRNA, carrying the amino acid methionine, binds to the start codon in the mRNA’s ribosome-binding site.
- Elongation: During elongation, the ribosome moves along the mRNA, adding amino acids to the growing polypeptide chain. The process involves three main steps:a. Codon recognition: Each codon in the mRNA is recognized by a complementary anticodon on a specific tRNA molecule. The tRNA carries the corresponding amino acid that matches the codon. The ribosome ensures the correct matching of codons and anticodons.b. Peptide bond formation: The ribosome catalyzes the formation of a peptide bond between the amino acid carried by the incoming tRNA and the growing polypeptide chain. The amino acid is transferred from the tRNA to the polypeptide chain.c. Translocation: After peptide bond formation, the ribosome moves one codon along the mRNA in the 5′ to 3′ direction. This movement requires energy in the form of GTP (guanosine triphosphate). The tRNA that was previously in the A site (aminoacyl site) is now in the P site (peptidyl site), and the empty tRNA is in the E site (exit site). This step prepares the ribosome for the next cycle of elongation.
- Termination: Termination occurs when the ribosome reaches a stop codon (UAA, UAG, or UGA) in the mRNA. No tRNA molecules carry amino acids that correspond to stop codons. Instead, release factors bind to the ribosome, causing the release of the completed polypeptide chain. The ribosome subunits dissociate from the mRNA, and the protein synthesis machinery resets for the next round of translation.
- Post-translational modifications: After translation, the newly synthesized polypeptide may undergo various modifications to become a functional protein. These modifications can include folding, cleavage of signal sequences or additional amino acids, phosphorylation, acetylation, glycosylation, or association with other proteins. These modifications are essential for the proper structure and function of the protein.
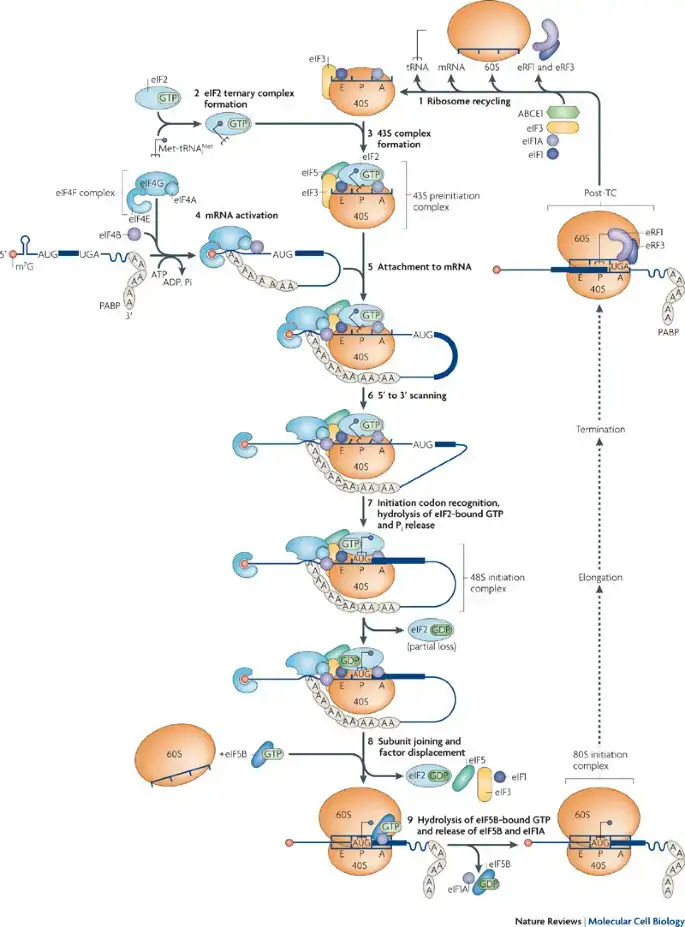
It’s important to note that translation in eukaryotic cells is more complex than in prokaryotes due to the presence of additional regulatory factors, compartmentalization of cellular processes, and post-transcriptional modifications. The process is tightly regulated and subject to various checkpoints to ensure accurate protein synthesis.
Regulation of Translation In Eukaryotes
1. Protein Synthesis Regulation in Eukaryotes through Changes in Genes
Regulation through changes in genes plays a vital role in controlling gene expression and protein synthesis. Here are some examples of how genes can be regulated through changes in their structure or activity:
- Gene loss or partial loss: During cellular differentiation, certain cells may undergo gene loss or partial loss, resulting in the absence or reduced expression of specific genes. This process allows cells to specialize and perform specific functions. For example, during the maturation of red blood cells, they lose their nuclei along with many genes, including those responsible for protein synthesis. This ensures that mature red blood cells focus solely on their oxygen-carrying function.
- Gene amplification: In some cases, genes can be amplified, leading to an increased number of copies within the genome. This amplification can result in higher production levels of the encoded protein. Methotrexate, a drug used in cancer treatment, induces the amplification of the dihydrofolate reductase gene. The increased gene copies enable cells to produce more of the enzyme, conferring resistance to the drug.
- DNA segment movement and rearrangement: Segments of DNA can move from one location to another within the genome, leading to different arrangements of genetic material. This rearrangement can result in altered gene expression and the production of different proteins. For instance, during the development of antibodies, a process called V(D)J recombination occurs, where different segments of the antibody gene rearrange to generate a diverse repertoire of antibodies capable of recognizing various antigens.
- DNA base modifications: Modifications to the bases within DNA can impact gene expression and protein synthesis. One common modification is methylation, which involves adding a methyl group to the DNA molecule. Methylation often occurs at specific sites known as CpG islands, typically found within promoter regions. When cytosine is methylated at the 5th position, it can inhibit gene transcription. The greater the extent of methylation, the more likely the gene is to be silenced or expressed at lower levels.
These regulatory mechanisms highlight the dynamic nature of gene expression. Through gene loss, amplification, rearrangement, and base modifications, cells can finely tune protein production and respond to specific developmental, environmental, or pathological cues. Understanding these regulatory processes is crucial for comprehending normal cellular function as well as the mechanisms underlying various diseases.
2. Protein Synthesis Regulation in Eukaryotes through Regulation of the level of transcription
The regulation of the level of transcription is a critical process that determines the amount of mRNA synthesized from a specific gene. It involves various mechanisms that control the accessibility of DNA to the transcriptional machinery. Here are some key factors and processes involved in the regulation of transcription levels:
- Histone-mediated regulation: Histones are small, basic proteins associated with DNA in eukaryotes. They play a crucial role in regulating gene expression by modulating the accessibility of DNA to the transcriptional machinery. In general, histones act as non-specific repressors, tightly packaging DNA into a structure known as heterochromatin, which results in the transcriptional inactivity of associated genes. Histone acetyltransferases (HATs) catalyze the acetylation of lysine residues on histones. Acetylation reduces the positive charge on histones, weakening their interaction with DNA and promoting a more open chromatin structure called euchromatin. This relaxed chromatin structure allows for increased accessibility of the DNA, facilitating transcription. Conversely, histone deacetylases (HDACs) remove the acetyl groups, promoting the reassociation of histones with DNA and leading to transcriptional repression.
- Heterochromatin and euchromatin: Heterochromatin represents tightly packed chromatin associated with inactive genes, while euchromatin refers to the more loosely packed chromatin associated with active genes. The balance between heterochromatin and euchromatin plays a critical role in transcriptional regulation. The level of compaction of chromatin determines the accessibility of the DNA to transcription factors and other regulatory proteins, influencing transcriptional activity.
- Positive regulation of gene expression: Certain genes require positive mechanisms to stimulate their transcription. This typically involves the binding of inducers to specific protein receptors. For example, steroid hormones can enter cells and interact with nuclear receptors. The hormone-receptor complex then binds to specific regulatory regions of the chromatin, activating the transcription of target genes.
- Alternative promoters: Some genes have multiple promoters, allowing for the generation of different transcripts and protein isoforms. The choice of which promoter is used can vary under different physiological conditions or in different cell types. This provides flexibility in gene expression and allows for the production of distinct protein products with specific functions.
The regulation of transcription levels is a complex process involving the interplay of various factors, including histone modifications, chromatin structure, inducers, and the usage of alternative promoters. These mechanisms ensure precise control over gene expression, allowing cells to respond to internal and external cues and maintain cellular homeostasis.
3. Protein Synthesis Regulation in Eukaryotes by Chromatin Remodeling
Regulation by chromatin remodeling is a crucial mechanism that modulates gene expression by altering the structure and accessibility of chromatin. Here are some key aspects of chromatin remodeling:
- Nucleosome displacement: Chromatin is organized into nucleosomes, which consist of DNA wrapped around histone proteins. Nucleosome displacement is a process that involves the movement or removal of nucleosomes, allowing the transcriptional machinery access to the underlying DNA. This displacement is essential for transcription to occur effectively.
- ATP-driven chromatin remodeling complexes: Chromatin remodeling complexes are multi-protein complexes that utilize ATP as an energy source to remodel chromatin structure. These complexes can recognize specific histone modifications associated with active chromatin, such as acetylation. The bromodomains present on proteins within the chromatin remodeling complex recognize and bind to acetylated histones. Once bound, the ATPase subunits within the complex utilize ATP hydrolysis to induce conformational changes, leading to the repositioning or eviction of nucleosomes. This chromatin remodeling activity creates open regions of DNA, allowing transcription factors and the transcriptional machinery to access the gene for transcription.
- Histone acetylase activity: Transcription factors involved in gene activation often recruit histone acetyltransferases (HATs) to the chromatin region that needs to be transcribed. Histone acetylation catalyzed by HATs neutralizes the positive charge of histones, weakening their interaction with DNA and promoting a more open chromatin structure. This modification facilitates the removal of histones from the DNA, making the gene region more accessible to the transcription apparatus. The binding of transcription factors, along with the acetylation of histones, synergistically contributes to the activation of gene transcription.
In summary, chromatin remodeling plays a crucial role in the regulation of gene expression. Through ATP-driven chromatin remodeling complexes and histone acetylase activity, chromatin structure is modified, leading to nucleosome displacement, increased accessibility of DNA, and facilitation of transcription. These processes allow for dynamic control of gene expression, enabling cells to respond to developmental cues, environmental changes, and various physiological signals.
4. Protein Synthesis Regulation in Eukaryotes during processing and transport of mRNA
Regulation during the processing and transport of mRNA is an important aspect of gene expression control. Here are some key regulatory mechanisms involved:
- Capping, polyadenylation, and splicing: These processes occur during mRNA processing and can significantly impact the final protein product. Regulatory mechanisms can influence these steps to alter the amino acid sequence or the quantity of the protein produced. Alternative splicing allows different combinations of exons within a gene to be included or excluded in the mature mRNA, resulting in the production of multiple protein isoforms from a single gene. Similarly, alternative polyadenylation sites can be used, leading to the generation of different mRNA isoforms with variable lengths of the 3′ untranslated region (UTR). These variations can affect translation efficiency, stability, and localization of the mRNA.
- mRNA degradation: The rate of mRNA degradation is tightly regulated and contributes to the control of gene expression. Nucleases can degrade mRNA molecules both in the nucleus, before their transport to the cytoplasm, and in the cytoplasm itself. The stability of an mRNA molecule can be influenced by specific sequences, secondary structures, or binding of regulatory proteins or small non-coding RNAs.
- RNA editing: RNA editing involves post-transcriptional alterations of mRNA, where specific bases are modified. This process can lead to changes in the coding sequence, resulting in an altered amino acid sequence in the translated protein. RNA editing can be mediated by enzymes that modify individual nucleotides, such as adenosine deaminases.
- Small RNA molecules: Small RNA molecules, such as microRNAs (miRNAs) and small interfering RNAs (siRNAs), play a crucial role in regulating gene expression. miRNAs, derived from precursor molecules, bind to target mRNAs and either inhibit their translation or induce their degradation. This gene silencing mechanism allows for the fine-tuning of gene expression by suppressing the production of specific proteins. miRNA genes are located throughout the genome, with some even present within introns of the genes they regulate. These small RNA molecules are synthesized in the nucleus and processed to form active miRNAs that can target and downregulate specific mRNAs.
Regulation during mRNA processing and transport adds an additional layer of control to gene expression. Through alternative splicing, polyadenylation, RNA editing, mRNA degradation, and small RNA-mediated gene silencing, cells can modulate the composition, stability, and translation efficiency of mRNA molecules, thereby influencing protein synthesis and cellular processes. These mechanisms play crucial roles in development, tissue specificity, and response to various stimuli.
5. Protein Synthesis Regulation in Eukaryotes at the translational level
Regulation at the translational level is a fundamental process that controls the rate of protein synthesis. Here are some key regulatory mechanisms involved in translational regulation:
- Initiation and elongation reactions: Regulation can occur during the initiation or elongation phases of protein synthesis. During translation initiation, specific factors are required to assemble the ribosome on the mRNA and initiate protein synthesis. Regulatory signals can modulate the activity of these initiation factors, influencing the efficiency of translation.
- Heme and eIF-2 regulation: Heme, a molecule involved in oxygen transport, can regulate the synthesis of globin, a component of hemoglobin. It achieves this by preventing the phosphorylation and subsequent inactivation of eIF-2, a translation initiation factor. When eIF-2 is active, it facilitates the recruitment of ribosomes to initiate protein synthesis. By maintaining eIF-2 in its active form, heme stimulates the synthesis of globin and ensures the proper production of hemoglobin.
- Interferon and eIF-2 regulation: Interferons, which are signaling molecules involved in immune responses, can regulate translation through the phosphorylation of eIF-2. When interferon signaling is activated, it leads to the phosphorylation of eIF-2, which inhibits translation initiation. This regulation serves as a mechanism to dampen protein synthesis during viral infections or cellular stress, helping to restrict viral replication and promote cellular defense mechanisms.
- Iron-response elements (IREs) in mRNA: Iron-responsive elements are specific RNA sequences found in the mRNA molecules of certain genes involved in iron homeostasis. For example, the mRNA for ferritin, an iron storage protein, and the transferrin receptor, which mediates iron uptake, contain IREs. These elements provide a regulatory mechanism that responds to iron levels in the cell. When iron levels are high, IREs in the ferritin mRNA promote translation, allowing for increased synthesis of ferritin to store excess iron. Conversely, IREs in the transferrin receptor mRNA destabilize the mRNA, reducing its translation and thereby decreasing iron uptake when iron levels are sufficient.
These examples highlight the diverse strategies employed to regulate translation. By modulating the activity of translation initiation factors, such as eIF-2, or by utilizing regulatory elements in mRNA molecules, cells can finely tune protein synthesis in response to specific signals, metabolic conditions, or cellular needs. Translational regulation allows for rapid and dynamic control of protein production, playing a crucial role in cellular processes, development, and adaptation to changing environments.
Significance of translational control in eukaryotic cells:
Translational control, which refers to the regulation of protein synthesis at the level of translation, plays a crucial role in diverse biological processes and has significant implications for cellular function and human health. Here are some key areas where translational control holds particular significance:
- Gene regulation during nutrient deprivation and stress: Translational control allows cells to quickly respond to changes in nutrient availability or stressful conditions. By regulating the translation of specific mRNAs, cells can prioritize the synthesis of proteins that are essential for survival and adaptation under adverse conditions. This ensures efficient resource utilization and promotes cell survival during times of limited resources or external challenges.
- Development and differentiation: Translational control is vital for proper development and cellular differentiation. During embryonic development, precise regulation of protein synthesis is required for the coordinated growth and differentiation of different cell types and tissues. Translational control mechanisms play a role in establishing and maintaining cell identity, guiding cell fate decisions, and orchestrating complex developmental processes.
- Nervous system function: Translational control is particularly important in the nervous system, where precise spatial and temporal regulation of protein synthesis is critical for processes such as synaptic plasticity, learning, and memory formation. Localized translation at synapses enables rapid and targeted protein synthesis in response to neuronal activity, allowing for the efficient modulation of synaptic strength and the establishment of long-lasting neural connections.
- Aging: Translational control has been implicated in the aging process and age-related diseases. Dysregulation of translation can lead to altered protein homeostasis, impaired cellular functions, and increased susceptibility to age-associated pathologies. Maintaining proper translational control is crucial for healthy aging and the maintenance of tissue and organ function.
- Disease: Translational control plays a role in various diseases, including cancer, neurodegenerative disorders, and metabolic disorders. Aberrant regulation of translation can result in the overproduction or underproduction of specific proteins, contributing to disease progression. Targeting translational control pathways has emerged as a potential therapeutic strategy for a range of diseases, offering the possibility of modulating protein synthesis and restoring cellular balance.
FAQ
What is protein synthesis regulation?
Protein synthesis regulation refers to the control and modulation of the process by which proteins are produced in a cell. It involves various mechanisms that influence the rate and efficiency of translation, which is the process of synthesizing proteins from messenger RNA (mRNA) molecules.
What are the key mechanisms involved in regulating protein synthesis in eukaryotic cells?
Key mechanisms of protein synthesis regulation in eukaryotic cells include transcriptional control, post-transcriptional modifications, translational control, and mRNA degradation. These mechanisms involve the actions of transcription factors, RNA-binding proteins, ribosomes, and regulatory molecules that modulate the initiation, elongation, and termination steps of translation.
How are gene expression and protein synthesis connected?
Gene expression refers to the overall process by which genetic information encoded in DNA is used to produce functional proteins. Protein synthesis is a crucial step in gene expression where the information encoded in mRNA molecules is translated into protein molecules. Therefore, protein synthesis is an integral part of gene expression.
What are the roles of transcription factors in protein synthesis regulation?
Transcription factors are proteins that bind to specific DNA sequences and regulate the initiation and rate of transcription. They can influence protein synthesis by controlling the production of mRNA molecules, which are then used as templates for protein synthesis. Transcription factors can either promote or inhibit the transcription of specific genes, thereby regulating the availability of mRNA for translation.
How do post-transcriptional modifications affect protein synthesis?
Post-transcriptional modifications, such as mRNA capping, polyadenylation, and splicing, can impact protein synthesis. These modifications can alter the stability, structure, and translational efficiency of mRNA molecules. For example, alternative splicing can generate different mRNA isoforms, leading to the production of distinct protein variants from a single gene.
What is the role of translation initiation factors in protein synthesis regulation?
Translation initiation factors are proteins that facilitate the assembly of ribosomes on mRNA molecules and promote the initiation of protein synthesis. They play a crucial role in regulating the rate of translation and can be targeted by various regulatory mechanisms to control protein synthesis. Factors like eIF-2 (eukaryotic initiation factor 2) can be phosphorylated or dephosphorylated, affecting their activity and modulating translation initiation.
How does mRNA degradation contribute to protein synthesis control?
mRNA degradation is an essential mechanism for controlling protein synthesis. The degradation of mRNA molecules can be influenced by various factors, including RNA-binding proteins and ribonucleases. The rate of mRNA degradation determines the availability and lifespan of mRNA templates for translation, thereby regulating protein synthesis levels.
What is the significance of alternative splicing in protein synthesis regulation?
Alternative splicing is a process where different combinations of exons within pre-mRNA can be joined together, leading to the generation of multiple mRNA isoforms from a single gene. Alternative splicing provides cells with the ability to produce different protein variants from a single gene, expanding the proteome diversity and allowing for fine-tuned regulation of protein synthesis.
How do small non-coding RNAs, such as miRNAs, impact protein synthesis regulation?
Small non-coding RNAs, such as microRNAs (miRNAs), play a crucial role in post-transcriptional gene regulation. miRNAs can bind to complementary sequences on mRNA molecules, resulting in the inhibition of translation or degradation of target mRNAs. By targeting specific mRNAs, miRNAs can fine-tune protein synthesis levels and contribute to the regulation of various cellular processes.
How do cellular stress and environmental factors influence protein synthesis regulation?
Cellular stress and environmental factors can modulate protein synthesis regulation. For example, nutrient deprivation, oxidative stress, and viral infections can trigger signaling pathways that alter the activity of translation initiation factors, leading to global changes in protein synthesis rates. Additionally, specific stress-responsive transcription factors can regulate the expression of genes involved in protein synthesis and cellular adaptation to stress conditions.
References
- Sonenberg, N., & Hinnebusch, A. G. (2009). Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell, 136(4), 731-745.
- Jackson, R. J., Hellen, C. U., & Pestova, T. V. (2010). The mechanism of eukaryotic translation initiation and principles of its regulation. Nature Reviews Molecular Cell Biology, 11(2), 113-127.
- Gebauer, F., & Hentze, M. W. (2004). Molecular mechanisms of translational control. Nature Reviews Molecular Cell Biology, 5(10), 827-835.
- Hinnebusch, A. G. (2014). The scanning mechanism of eukaryotic translation initiation. Annual Review of Biochemistry, 83, 779-812.
- Gebauer, F., & Preiss, T. (2019). Hentze, MW. From cis-regulatory elements to complex RNPs and back. Cold Spring Harbor Perspectives in Biology, 11(6), a032814.
- Sonenberg, N., & Dever, T. E. (2003). Eukaryotic translation initiation factors and regulators. Current Opinion in Structural Biology, 13(1), 56-63.
- Jivotovskaya, A. V., & Valasek, L. S. (2009). RNA-protein interactions in the eukaryotic ribosome: the structures and functions of protein-binding sites. Wiley Interdisciplinary Reviews: RNA, 1(1), 155-168.
- Sen, N. D., Zhou, F., & Ingolia, N. T. (2020). Genome-wide analysis of translational efficiency reveals distinct but overlapping functions of yeast DEAD-box RNA helicases Ded1 and eIF4A. Genome Research, 30(3), 437-449.
- Hinnebusch, A. G., Ivanov, I. P., & Sonenberg, N. (2016). Translational control by 5′-untranslated regions of eukaryotic mRNAs. Science, 352(6292), 1413-1416.
- von der Haar, T. (2008). A quantitative estimation of the global translational activity in logarithmically growing yeast cells. BMC Systems Biology, 2(1), 87.