- Lipopolysaccharide (LPS) is the main component of the outer membrane of Gram-negative bacteria.
- Lipopolysaccharide is found in the outermost membrane layer and is in noncapsulated strains visible on the cell’s surface.
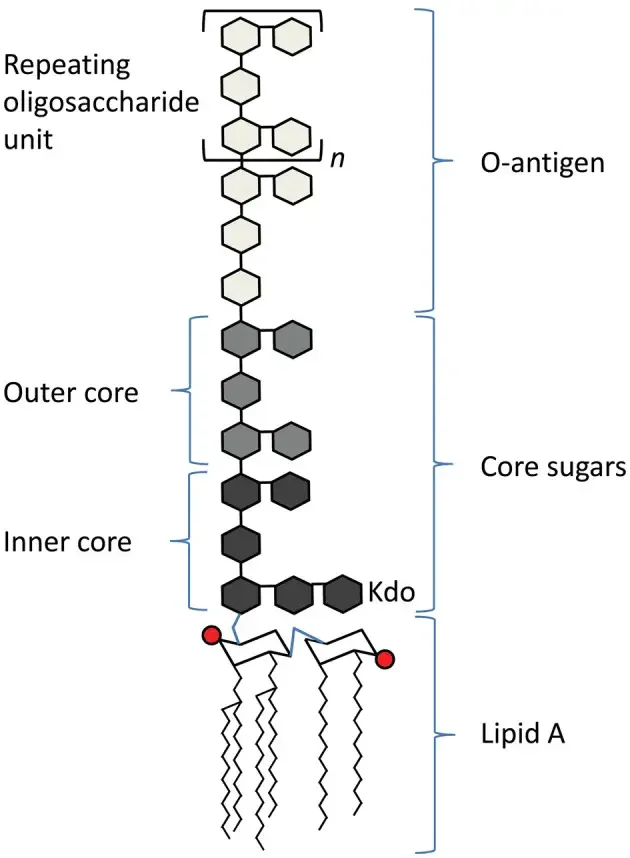
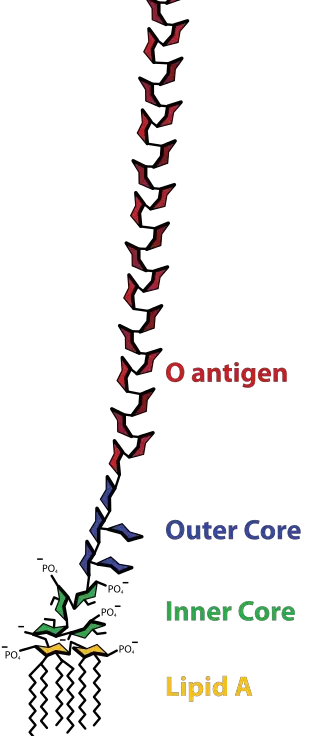
- Lipopolysaccharides (LPS) are huge molecules that comprise the lipid and an O-antigen-based polysaccharide an outer core and an inner core that are joined via covalent bonds.
- The term “lipooligosaccharide (“LOS”) refers to a used to describe an bacterial form with a lower molecular weight.
- In the present, the term “endotoxin” is usually used to refer the LPS but there are some endotoxins (in the sense of toxins that are less secreted) which aren’t connected to LPS for instance, the delta endotoxin proteins made by Bacillus Thuringiensis.
- The toxic nature that is a result of LPS was first identified and named endotoxin by Richard Friedrich Johannes Pfeiffer, who distinguished between exotoxins which were classified as a toxins that bacteria release to the environment and endotoxins which he considered to be a toxins stored “within” the bacterial cell and released only upon destruction of the cell wall.
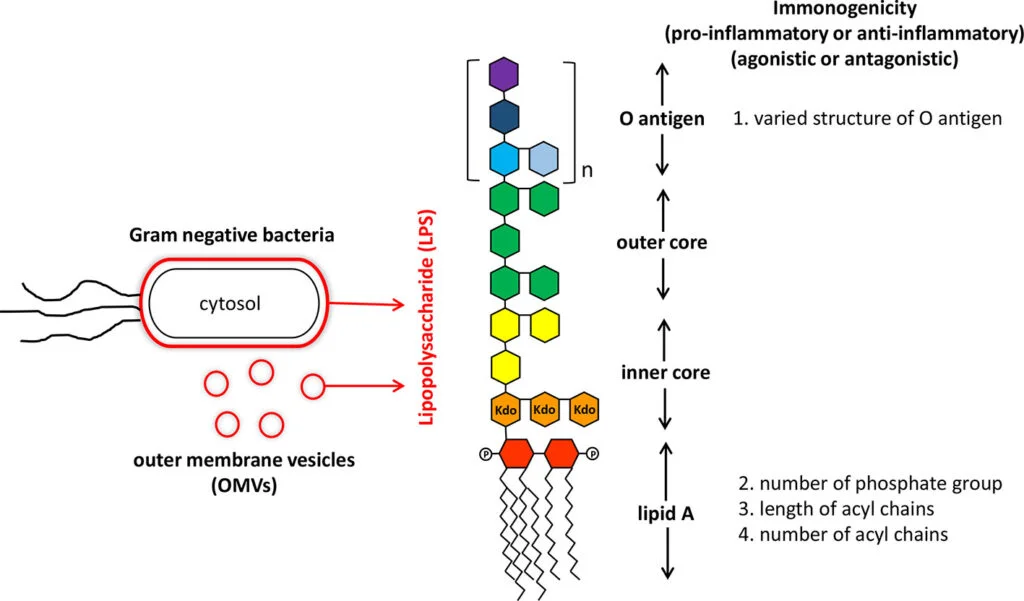
Structure and Composition of Lipopolysaccharide (LPS)
It is composed of three components like;
- O antigen (or O polysaccharide)
- Core oligosaccharide
- Lipid A
O-antigen
- A polymer of glycans that is repeated in an LPS is called O antigen, O antigen. O polysaccharide or O chain of bacteria’s side.
- Its O antigen is bound to the oligosaccharide’s core and forms the outermost region that is part of the LPS molecule.
- Its composition in the O chain differs according to the strain. In the case of E. coli, for instance there are more than 160 distinct O antigen structures created by various E. bacteria strains.
- In the absence or presence or absence of O chains determines if an LPS is deemed rough or smooth.
- O-chains of full length would make the LPS smooth and smooth, while the removal or reduction of O chains makes your LPS rough.
- The bacteria with rough LPS generally have cell membranes that are more permeabil to hydrophobic antibiotics since rough LPS is more hydrophobic.
- O antigen is visible on the outside that the cells of bacterium and as a result is a potential source of recognition by antibodies from the host.
Core
- The core domain always contains an oligosaccharide component that attaches directly to lipid A and commonly contains sugars such as heptose and 3-Deoxy-D-manno-oct-2-ulosonic acid (also known as KDO, keto-deoxyoctulosonate).
- The LPS the cores of a variety of bacteria also contain non-carbohydrate constituents including amino acids, phosphate and ethanolamine substitutes.
Structure of Core or Core oligosaccharide (or Core-OS)
The core domain always contains an oligosaccharide component which attaches directly to lipid A and commonly contains sugars such as heptose and 3-deoxy-D-mannooctulosonic acid (also known as KDO or keto-deoxyoctulosonate). It is also known as keto-deoxyoctulosonate. LPS Cores of a variety of bacteria also have non-carbohydrate elements like amino acids, phosphate and ethanolamine substitutes.
- Inner core: “base” of the inner core is composed of 1-3 KDO residues. The final KDO is usually modified by an ethanolamine or phosphate group. From the KDOs there are attached two Heptoses (i.e. L-glycero-D-mannoheptulose) that are usually phosphorylated. They KDO and heptoses form”the “inner core”. The ketosidic bond that connects KDO and the lipid A (α2-6) is especially prone to acid cutting. LPS researchers utilize the weak acid treatment to divide the polysaccharide and lipid parts of LPS. A LPS is a molecule with just a lipid A, and its internal center (or less. For an example, see) is known as “deep-rough LPS”.
- Outer core: This layer is comprised of hexose residues which attach to the last heptose residue within the core. The most frequent hexoses located within the core’s outer layer comprise D-glucose (D-mannose), D-glucose ( D-galactose, and so on.. It is common to find 3 or more hexoses that are bound to β1→3, with the O antigen being linked with the 3rd hexose. Other hexoses are typically connected to the outer core and branching from the principal oligomer. LPS that comprise lipid A and a full core of oligosaccharide (inner as well as outer) is called “rough LPS.”
Biosynthesis
The enzymes involved in the process of core oligosaccharide production are shared by Escherichia coli as well as Salmonella. Pseudomonas aeruginosa is a distinctive enzymes.
Lipid A
- Lipid A is normal conditions it is a phosphorylated glucosamine disaccharide embellished with various fatty acids.
- These hydrophobic chains of fatty acids secure the LPS to the membrane of the bacterial and the remainder of the LPS extends away from the cell’s surface.
- The domain of lipid A is responsible for a large portion of the toxicity of Gram-negative bacteria.
- In the event that bacterial cells are lysed through the immune system fragments of the membrane that contain liquid A can be released to the blood circulation which can cause fever, diarrhea and possibly fatal endotoxic shock (also known as Septic shock).
- Lipid A moiety Lipid A moiety of the LPS is a highly conserved part that is a part of LPS.
- But Lipid A structure differs between bacterial species. Lipid A structure is a signpost for an overall immunity activation of the host.
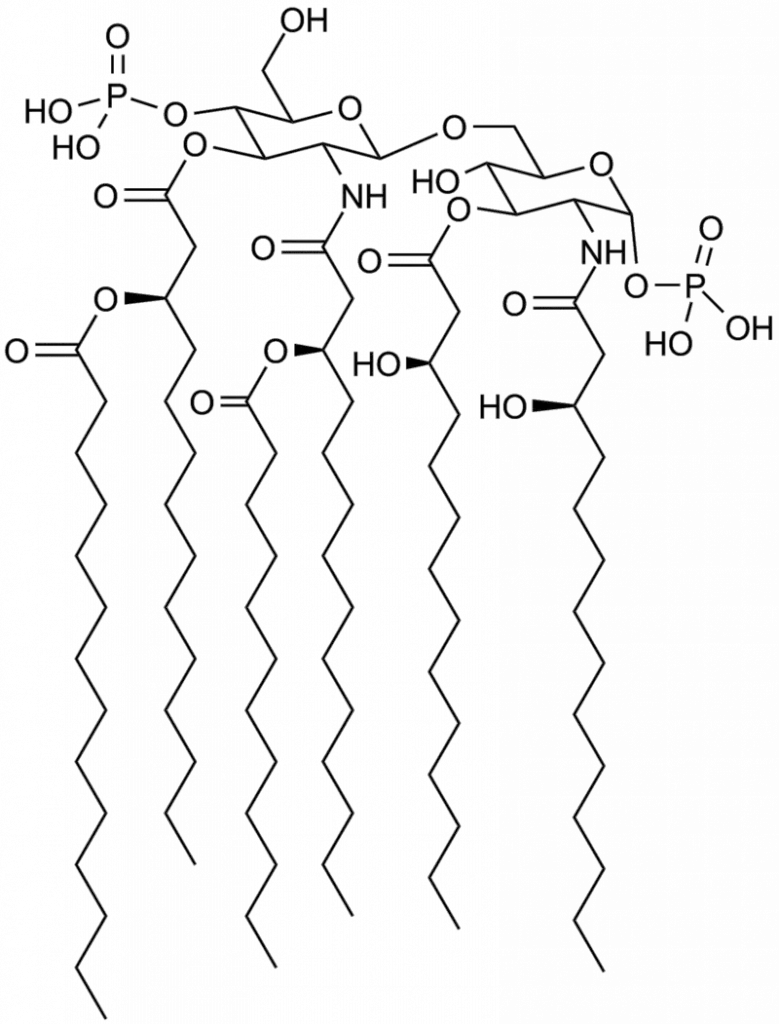
Structure of Lipid A
- Lipid A is composed consisting of two glucosamine (carbohydrate/sugar) elements, connected by the β(1→6) linkage which has attached chains of acyl (“fatty acid”) and typically having one phosphate group in each carbohydrate.
- The most effective immune activating lipid structure is believed to have six acyl chains.
- Four acyl chains that are directly attached to the sugars glucosamine comprise betahydroxy chains that are typically between 10 and 16 carbons long.
- Two other acyl chains are usually linked to the beta hydroxy group.
- E. coli lipid A is an example. It typically contains four C14 chain hydroxy acyls connected to sugars, along with the one C14 along with one C12 connected to betahydroxy groups.
Biosynthesis of of Lipid A
The enzymes that are involved with Lipid A synthesis are conserved between Pseudomonas Aeruginosa Escherichia Coli, Bordetella bronchiseptica, and Salmonella.
Functions of Lipopolysaccharides (LPS) Layer
- The outer membrane acts as an impermeable barrier that helps stop the flow of vital enzymes, like the ones involved in the development of cell walls and cell wall growth, out of the periplasmic space. It also acts as a shield against external substances and enzymes that may harm the cell.
- Outer membranes allow smaller molecules, like monosaccharides, nucleotides, oligosaccharides and amino acids, through porin channels.
- Lipopolysaccharide is a known pyrogenic (responsible to cause fever) substance, which can also trigger an endotoxic shock. LPS stimulates macrophages and leads into the release of TNF alpha, IL1 – 1 and the IL-6. IL1 1 is the most important mediator of fever.
- Macrophage activation, as well as its products, can cause tissue injury.
- The endothelium is damaged by bradykinin-induced vasodilation causes shock.
- In the process, coagulation (DIC) is controlled by activated Hageman factor (coagulation factor XII).
References
- https://en.wikipedia.org/wiki/Lipopolysaccharide
- https://www.researchgate.net/figure/Schematic-of-the-basic-structure-of-lipopolysaccharide-LPS-consists-of-three-regions_fig1_235621135
- https://www.sigmaaldrich.com/IN/en/technical-documents/technical-article/research-and-disease-areas/cell-signaling/lipopolysaccharides
- https://microbeonline.com/lipopolysaccharide-lps-of-gram-negative-bacteria-characteristics-and-functions/