What is Gene frequencies?
Gene frequencies, also known as allele frequencies, refer to the relative proportions of different alleles at a particular locus or gene within a population. They represent the prevalence or abundance of specific alleles in relation to the total number of alleles in the gene pool.
Gene frequencies are typically expressed as a decimal or a percentage. For a diploid organism with two alleles at a given locus, the sum of the allele frequencies at that locus will always equal 1 or 100%.
For example, let’s consider a population of rabbits with a gene that determines fur color. The gene has two alleles: A (for brown fur) and a (for white fur). If the frequency of the A allele is 0.6 (or 60%) and the frequency of the a allele is 0.4 (or 40%), the gene frequencies can be expressed as follows:
- Frequency of allele A = 0.6 or 60%
- Frequency of allele a = 0.4 or 40%
It’s important to note that gene frequencies can change over time due to various factors such as mutation, gene flow, genetic drift, and natural selection. These changes in gene frequencies are the basis for understanding the processes of evolution and population genetics.
Definition of Gene frequencies
Gene frequencies, also known as allele frequencies, are the proportions or relative abundance of specific alleles at a particular gene locus within a population. They represent the frequency at which different versions of a gene (alleles) occur in a population and are typically expressed as decimals or percentages. Gene frequencies can change over time due to various evolutionary forces such as mutation, gene flow, genetic drift, and natural selection.
Genotype Frequency and Hardy-Weinberg Equilibrium
Genotype frequency and Hardy-Weinberg equilibrium are concepts that provide insights into the genetic composition of populations and how it remains stable over generations under certain conditions.
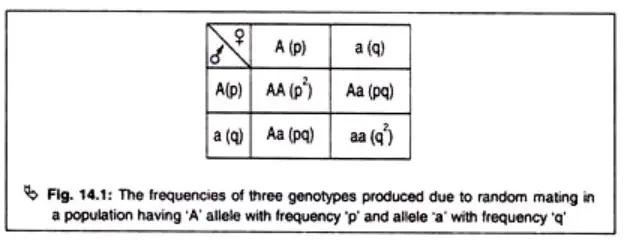
According to the Hardy-Weinberg law, in a randomly mating Mendelian population, the gene and genotype frequencies will remain constant if there is no selection, mutation, migration, or genetic drift. This equilibrium occurs when the population is not experiencing any evolutionary forces that can change the genetic makeup.
In terms of genotype frequencies, homozygotes (individuals with identical alleles, such as AA or aa) are produced through the union of gametes carrying the same alleles. The frequency of homozygotes can be calculated by multiplying the frequencies of the individual alleles. For example, the frequency of homozygotes with allele A (frequency p) mating with another homozygote with allele A will be p x p = p². Similarly, the frequency of homozygotes with allele a (frequency q) mating with another homozygote with allele a will be q x q = q².
Heterozygotes (individuals with two different alleles, such as Aa) are produced through the fusion of gametes carrying different alleles. The frequency of heterozygotes can be calculated by multiplying the frequencies of the two alleles. For example, the frequency of a male gamete carrying allele A (frequency p) fusing with a female gamete carrying allele a (frequency q) will be p x q = pq. This calculation can be applied in both directions, resulting in a total frequency of heterozygotes of 2pq.
The Hardy-Weinberg equation, (p + q)² = p² + 2pq + q² = 1, represents the frequencies of the two alleles in a population. The sum of the frequencies of alleles A and a (p + q) is always equal to 1. This equation allows for the calculation of gene frequencies based on observed phenotypic characteristics of homozygotes and heterozygotes.
The Hardy-Weinberg equilibrium can be used to determine the expected frequencies of genotypes in a population by comparing them to observed values. By applying the equation and comparing the expected and observed values, we can assess whether a population is in Hardy-Weinberg equilibrium.
Understanding genotype frequencies and Hardy-Weinberg equilibrium provides valuable insights into the genetic structure of populations and can help in studying the mechanisms of inheritance, estimating allele frequencies, and assessing the impact of evolutionary forces on gene frequencies.
Example 1: MN Blood Group of Humans The MN blood group in humans is controlled by two alleles, M and N. If we observe a population and find the following frequencies: MM = 0.2911, MN = 0.4984, and NN = 0.2121, we can calculate the expected genotype frequencies using the Hardy-Weinberg equation.
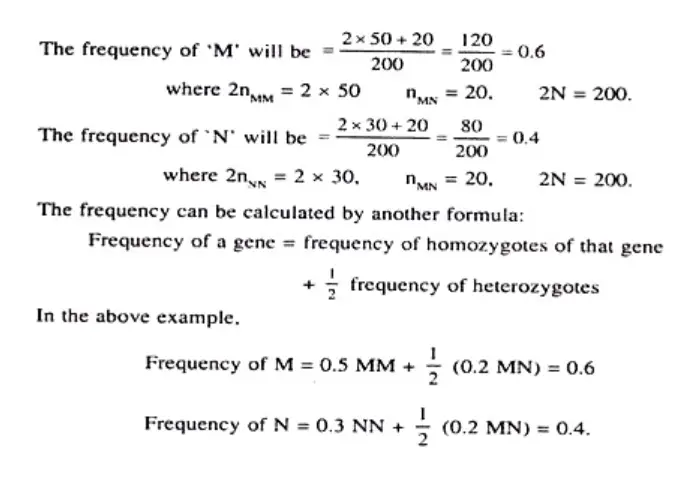
Using the equation (p + q)² = p² + 2pq + q² = 1, we can calculate the allele frequencies: p (frequency of M allele) = MM + 0.5MN = 0.2911 + (0.5 x 0.4984) = 0.5403 q (frequency of N allele) = NN + 0.5MN = 0.2121 + (0.5 x 0.4984) = 0.4597
Next, we can calculate the expected genotype frequencies: MM = p² = (0.5403)² ≈ 0.2919 MN = 2pq = 2 x 0.5403 x 0.4597 ≈ 0.4967 NN = q² = (0.4597)² ≈ 0.2114
Comparing the observed and expected genotype frequencies, we can determine whether the population is in Hardy-Weinberg equilibrium.
Example 2: Phenylketonuria (PKU) Frequency Phenylketonuria (PKU) is a genetic disorder that is controlled by a recessive allele. If we observe that 0.04% of a population is affected by PKU, we can use the Hardy-Weinberg equation to estimate the allele frequencies.
Let’s assume the frequency of the PKU allele is q² = 0.0004. Taking the square root of q², we can find q ≈ 0.02, which represents the frequency of the recessive allele. Since the dominant allele frequency is (1 – q) ≈ 0.98, we can estimate the number of carriers (heterozygotes) in the population using the equation 2pq.
2pq = 2 x 0.02 x 0.98 ≈ 0.0396
Thus, approximately 1.98% of the population are carriers of the PKU allele in the heterozygous condition.
These examples demonstrate how genotype frequencies can be calculated using the Hardy-Weinberg equation and how the concept of equilibrium helps assess the genetic stability of populations.
Factors Affecting Gene Frequency
Populations are dynamic entities that undergo changes over time. Various factors contribute to these changes, altering the genetic composition of populations and influencing gene frequencies. Understanding these factors is crucial for comprehending how populations evolve and adapt. Let’s explore some key factors that affect gene frequency:
- Mutations: Mutations introduce new alleles into a population. Although the rate of mutation is generally low, these rare occurrences contribute to genetic diversity. While some mutations may be beneficial and provide an evolutionary advantage, most mutations are either neutral or harmful. Natural selection plays a role in countering the effects of deleterious mutations by reducing their frequency.
- Migration: The movement of individuals between populations can significantly impact gene frequencies. When individuals migrate from one population to another, they bring their unique genetic makeup, introducing new alleles and potentially altering the gene pool. Migration can lead to genetic exchange, causing gene frequencies to change in both the source and recipient populations.
- Natural Selection: Natural selection acts as a powerful force in shaping gene frequencies. It favors certain genotypes that confer advantages in terms of survival and reproduction. Individuals with favorable traits are more likely to survive and pass on their genetic material to the next generation, leading to an increase in the frequency of beneficial alleles. Conversely, individuals with less advantageous traits may have reduced fitness, resulting in a decrease in the frequency of associated alleles.
- Random Genetic Drift: Random genetic drift refers to chance fluctuations in gene frequencies within a population. This factor is particularly significant in small populations, where random events can have a greater impact due to the limited number of individuals. Genetic drift can lead to the loss or fixation of alleles, resulting in genetic changes that are independent of selection pressure.
- Assortative Mating: Assortative mating occurs when individuals preferentially mate with others who share similar or dissimilar genotypes. This non-random mating can influence gene frequencies by affecting the frequency of homozygotes and heterozygotes. Positive assortative mating increases the frequency of homozygotes, while negative assortative mating increases the frequency of heterozygotes.
- Subpopulations and Inbreeding: Within larger populations, subpopulations may exist based on factors such as ethnicity, geographic location, or social structure. These subpopulations tend to mate more frequently within their local group, leading to inbreeding. Inbreeding increases the frequency of homozygotes and can result in a reduction of genetic diversity within the population.
Understanding these factors is essential for studying the dynamics of gene frequencies within populations. By examining the interplay between mutation, migration, selection, genetic drift, assortative mating, and inbreeding, scientists can gain insights into the processes that shape the genetic variation we observe in populations today.
Remember, the gene frequency changes occurring in populations are the driving force behind evolution and the remarkable diversity of life on our planet.
Measuring of Genotype Frequencies
Measuring genotype frequencies is an important aspect of population genetics that provides insights into the genetic makeup of a population. There are several methods to estimate genotype frequencies, including phenotype frequency and allele frequency approaches. Let’s explore these methods in more detail.
1. Phenotype Frequency Approach: One way to estimate genotype frequencies is by analyzing the frequencies of observed phenotypes in a population. Let’s consider an example of three blood groups: A, AB, and B, determined by two alleles, IA and IB, at a single locus. In a random sample of 1000 individuals, we find 210 individuals with blood group A, 450 with AB, and 340 with B.
To calculate the frequency of each blood group phenotype, we divide the number of individuals with each blood group by the total sample size. For instance, the frequency of blood group B would be 340/1000 = 0.34.
2. Allele Frequency Approach: Another method to estimate genotype frequencies is by first determining the gene frequencies of the alleles in the population. Let’s assume that our sample contains 210 individuals with AA genotype, 450 with AB genotype, and 340 with BB genotype.
To calculate the gene frequency of allele A, we need to determine the proportion of A alleles among all alleles at the AB locus in the sample. Since each individual carries two alleles, the total number of alleles in the sample is 1000 x 2 = 2000. Out of these, 210 + 210 + 450 = 870 alleles are A. Therefore, the frequency of the A allele is 870/2000 = 0.435. Similarly, the frequency of the B allele is 1130/2000 = 0.565.
We can represent the gene frequency of A as “p” and the frequency of B as “q.” It’s important to note that q = 1 – p and p = 1 – q. Thus, p + q = 1.
3. Predicting Genotype Frequencies: To predict genotype frequencies, certain assumptions need to be made, such as random mating in the population. Random mating implies that individuals mate without regard to their specific genotypes. In the case of blood groups, an individual can mate with another regardless of whether the mate has an AA, AB, or BB genotype.
Now, based on the assumption of random mating, we can calculate the probabilities of different genotypes. The probability of an A allele in the population is “p,” which is also the probability of a randomly chosen gamete carrying allele A. Similarly, the probability of a B allele is “q.”
Using these probabilities, we can predict genotype frequencies. For example, the frequency of the AA genotype is p2, the frequency of the AB genotype is 2pq, and the frequency of the BB genotype is q2. By substituting the values of p = 0.435 and q = 0.565 from our previous calculations, we find that the frequency of AA = (0.435)2 = 0.189, AB = 2 x 0.435 x 0.565 = 0.246, and BB = (0.565)2 = 0.319.
By applying these methods, scientists can estimate the frequencies of different genotypes in a population, providing valuable insights into the genetic structure and diversity of a population. These estimations form the foundation for understanding genetic inheritance patterns, population dynamics, and the forces that shape genetic variation.
Remember, measuring genotype frequencies allows us to delve into the intricate details of population genetics and uncover the fascinating genetic makeup of living organisms.
Common Mistakes to Avoid
When studying population genetics, it’s important to be aware of common mistakes that can lead to misconceptions or inaccurate interpretations. By avoiding these pitfalls, you can gain a clearer understanding of the complexities of genetic variation and evolution. Let’s explore some common mistakes to avoid:
- Trying to Find p First: One mistake often made by students is attempting to calculate the frequency of the dominant allele (p) by directly observing the population and taking the square root of the observed proportion. However, this approach does not work in typical recessive/dominant allele relationships. The presence of a dominant allele can mask the presence of a recessive allele. Therefore, simply calculating the square root of the observed frequency can overestimate the p allele frequency. Heterozygous individuals carrying the recessive allele should not be counted towards p.
- Relating Allele Frequency to Fitness: A common misconception is to assume that allele frequency is directly linked to the evolutionary fitness of a particular allele. However, the frequency of an allele does not determine its fitness. For instance, recessive traits that are deleterious can “hide” in a population, appearing at low levels. These traits may be present in heterozygous individuals and go unnoticed. Therefore, the frequency of an allele alone does not indicate its fitness or evolutionary advantage.
Moreover, beneficial mutations that confer a fitness advantage may have a low allele frequency initially. A new allele needs time to establish itself in a population by outcompeting other alleles and increasing its frequency. It must be continuously replicated across many generations. Therefore, even though a beneficial allele may have a low frequency, it does not imply that it is less fit. It may simply be in the early stages of spreading through the population.
By understanding these common mistakes, we can avoid misconceptions and develop a more accurate understanding of population genetics. It is crucial to consider the complexities of allele interactions, recessive traits, and the dynamic nature of evolutionary processes. Emphasizing a comprehensive and nuanced approach to studying population genetics will lead to a clearer interpretation of genetic variation and the mechanisms driving evolutionary change.
Remember, population genetics is a fascinating field that requires careful analysis and attention to detail. By avoiding these common mistakes, you can enhance your understanding and contribute to the advancement of our knowledge in the field of genetics.
How to Calculate Allele Frequency?
Calculating allele frequency is a fundamental step in population genetics, allowing us to understand the genetic composition of a population. To determine the frequency of a specific allele, scientists often employ the Hardy-Weinberg (HW) equation. Let’s explore the process of calculating allele frequency using this equation:
The Hardy-Weinberg equation is expressed as follows:
1 = p^2 + 2pq + q^2
Here, ‘p’ and ‘q’ represent the frequency of different alleles in the population. ‘p^2’ denotes the frequency of the homozygous dominant genotype, and ‘q^2’ represents the frequency of the homozygous recessive genotype.
To calculate the allele frequency, we can focus on the recessive phenotypes within the population. Recessive phenotypes are the result of having two recessive alleles. By determining the number of individuals with recessive phenotypes and dividing it by the total number of individuals in the population, we can estimate ‘q^2’.
Now, let’s take a closer look at an example to understand how to calculate the allele frequency of a specific allele:
Suppose we want to determine the frequency of allele ‘q’ in a population. We observe that there are 100 individuals with the recessive phenotype, which indicates the presence of two recessive alleles. The total population size is 500 individuals.
To calculate ‘q^2’, we divide the number of recessive individuals by the total population size:
q^2 = 100/500 = 0.2
Since ‘q^2’ represents the frequency of the homozygous recessive genotype, we can take the square root of ‘q^2’ to obtain the frequency of ‘q’:
q = √0.2 = 0.447
To find the frequency of the other allele (‘p’), we can use the equation p + q = 1:
p + 0.447 = 1
p = 1 – 0.447 = 0.553
Therefore, the frequency of allele ‘q’ is approximately 0.447, while the frequency of allele ‘p’ is approximately 0.553.
By utilizing the Hardy-Weinberg equation and analyzing the recessive phenotypes in a population, we can calculate the frequency of specific alleles. This information is vital for understanding the genetic diversity and dynamics of populations. Remember, accurate calculations of allele frequency contribute to our knowledge of population genetics and aid in various fields, such as evolutionary biology and medical research.
Allele Frequency Example
Understanding allele frequency is crucial in studying population genetics. Let’s dive into an example to grasp how allele frequencies can be calculated and interpreted.
Consider a simplified scenario where a population of rabbits has only two alleles, denoted as p and q. Additionally, we assume that there are no new mutations occurring in the population. In such a case, the sum of the allele frequencies of p and q must equal 1 since there are only two alleles present.
Let’s examine a hypothetical population of rabbits to illustrate the concept of allele frequency. In this population, a recessive allele determines whether a rabbit is white, while all other rabbits are black. Only rabbits with two copies of the recessive allele will exhibit the white phenotype.
Upon observing the population, we find that there are 16 white rabbits and 84 black rabbits. Since the white rabbits possess two copies of the recessive allele, their count directly represents q^2, the frequency of the homozygous recessive genotype.
In this case, the number of white rabbits is 16 out of the total population of 100 rabbits. Consequently, the frequency of the white rabbits, expressed as a percentage, is 16%, equivalent to 0.16. This value represents q^2, the frequency of the homozygous recessive genotype.
To determine the allele frequency of q (the white allele), we take the square root of q^2:
√0.16 = 0.4
Hence, the allele frequency of q (the white allele) is 0.4, corresponding to 40%.
To find the allele frequency of p (the black allele), we can simply subtract the frequency of q from 1. In this simplified scenario, where p and q are the only alleles and account for the entire set of alleles, p represents the remaining portion:
1 – 0.4 = 0.6
Therefore, the allele frequency of p (the black allele) is 0.6, accounting for 60% of the alleles.
In this example, we utilized the observed phenotypes of white and black rabbits to calculate the respective allele frequencies. Understanding allele frequencies provides insights into the genetic composition and diversity of populations. By applying this knowledge to real-world scenarios, scientists can unravel the intricate workings of evolution and make significant contributions to fields such as genetics and conservation biology.
Calculation of Gene Frequencies for Sex-Linked Genes
Understanding the frequencies of alleles in sex-linked genes requires a slightly modified approach due to the unique arrangement of sex chromosomes in different sexes. However, similar techniques can be employed to calculate gene frequencies for sex-linked traits.
In human populations, as well as in species like Drosophila, males are heterogametic sexes and possess only one X chromosome. Consequently, they cannot exhibit a binomial distribution for the random combination of pairs of sex-linked genes as females do. Nevertheless, we can still determine the equilibrium distribution of genotypes for sex-linked traits using a modified approach.
For a sex-linked trait, let’s designate the normal allele as R and the recessive allele as r. In the human population, approximately 8% of males are color-blind, which is a sex-linked recessive trait. This information tells us that the frequency of the gene r (q) is 0.08, and the frequency of its normal allele R (p) is 0.92.
To calculate the frequency of color-blind females, we square the frequency of the recessive allele:
q^2 = 0.0064
This value represents the expected frequency of color-blind females. Interestingly, this aligns with what is observed in real-world populations.
On the other hand, sex-linked dominant traits can also be analyzed using a similar approach. Let’s consider the example of normal color vision, which is a sex-linked dominant trait. Assuming the frequency of the normal allele R is 0.92 (p), we can calculate the incidence of normal females using the formula:
p^2 + 2pq = 0.9936
This value represents the expected frequency of normal females in the population.
Calculating gene frequencies for sex-linked genes allows us to understand the distribution of traits and their inheritance patterns. By applying these calculations, researchers can gain insights into the prevalence and impact of sex-linked traits within populations. This knowledge contributes to our understanding of genetics and helps inform various fields, including medicine and evolutionary biology.
FAQ
What are gene frequencies?
Gene frequencies refer to the relative frequencies of different alleles (alternative forms of a gene) in a population. They indicate how common or rare specific gene variants are within a group.
How are gene frequencies calculated?
Gene frequencies can be calculated by dividing the number of individuals in a population with a specific allele by the total number of individuals in the population.
What factors influence gene frequencies?
Gene frequencies can be influenced by several factors, including natural selection, genetic drift, mutation, migration, and assortative mating.
What is the Hardy-Weinberg equilibrium?
The Hardy-Weinberg equilibrium is a principle in population genetics that describes the distribution of gene frequencies in a non-evolving population. It assumes that gene frequencies remain constant from generation to generation in the absence of evolutionary forces.
How is the Hardy-Weinberg equilibrium equation used to calculate gene frequencies?
The Hardy-Weinberg equation (p^2 + 2pq + q^2 = 1) is used to calculate gene frequencies in a population. Here, p represents the frequency of one allele, q represents the frequency of the alternative allele, and p^2 and q^2 represent the frequencies of the two homozygous genotypes, while 2pq represents the frequency of the heterozygous genotype.
How can gene frequencies change due to natural selection?
Natural selection can change gene frequencies by favoring individuals with certain alleles that increase their fitness, thereby increasing the frequency of those alleles in subsequent generations.
What is genetic drift, and how does it affect gene frequencies?
Genetic drift is the random fluctuation of gene frequencies in a population over time due to chance events. It is more pronounced in small populations and can lead to the loss or fixation of alleles, resulting in changes to gene frequencies.
What is the role of mutation in altering gene frequencies?
Mutation introduces new genetic variation into a population. If a mutation provides a selective advantage, it can increase in frequency over time and alter gene frequencies.
How does migration impact gene frequencies?
Migration, the movement of individuals between populations, can introduce new alleles into a population or remove existing ones. It can lead to changes in gene frequencies as individuals with different allele frequencies join or leave a population.
Can assortative mating affect gene frequencies?
Assortative mating refers to the tendency of individuals to mate with those who have similar phenotypes. If assortative mating occurs based on certain genetic traits, it can lead to changes in gene frequencies by increasing the frequency of specific alleles in the population.