Dot blot is a blotting technique used for the detection of proteins. There are different blotting techniques such as Southern blotting, western blotting, and Northern blotting, etc.
What is Dot Blot?
- Dot Blot is a simplified technique of western blotting, which is mainly used for the detection of proteins.
- During the dot blotting, the electrophoresis of the protein samples not performed instead they are directly applied on a membrane in a single spot, and the blotting method is conducted.
- This method does not offer any information on the size of the target protein.
- It is considered as the most important technique in research and diagnostic laboratories used for the detection of known protein in a biological sample.
- The dot blotting knew as the slot blot technique.
Aim
- To understand the principle of Dot Blotting.
- Use of Dot blotting for diagnostic tests.
- To establish the importance of Dot blotting in identifying the protein of interest.
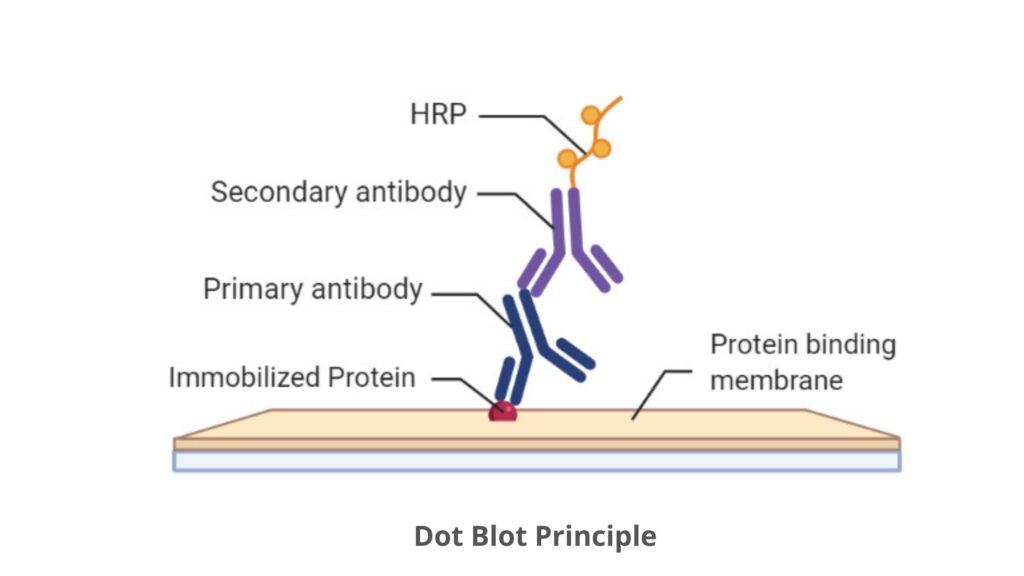
Dot Blot Principle
- The main characteristic feature of dot blot is the use of immunodetection techniques for the detection of a specific protein, for example, a protein marker for a disease.
- After the immobilization of proteins on a protein binding membrane (nitrocellulose or PVDF (polyvinylidene fluoride)) they are probed with a primary antibody, which is specific for the protein of interest.
- Once bound the antibody is visualized, either with a specific tag coupled to the primary antibody or with a secondary antibody.
- The secondary antibody helps to recognize the constant domain of immunoglobulin G and is species-specific. For example, if the primary antibody is a mouse antibody, the secondary antibody applied will identify all mouse antibodies.
- If a secondary antibody is applied then this will carry the tag that will help in the visualization of the protein.
- The enzymes are the most common tags used in Dot blot technique. These are catalyzed a substrate which results in the formation of either light that is recognized with radiography film, or color that is visualized on the membrane. Some examples of enzymes are horseradish peroxidase (HRP) and alkaline phosphatase (AP).
- An additional crucial step is performed in dot blot techniques is known as the blocking step. This step mainly increase the specificity of the Dot blot technique by preventing non‐specific interactions.
- If the membranes are not blocked then the antibodies can stick to non‐specific proteins due to their charge. To prevent this, the membrane is placed in a protein mixture and the proteins block the charges that would attract the antibodies.
- Different blocking against is used such as dried milk powder, bovine serum albumin and casein.
- Though, modern blocking agents apply synthetic and/or nonanimal proteins to prevent any cross-reaction with the animal antibodies. An example of a non-animal blocker is the provided NAP‐Blocker™.
Primary Antibodies
- On exposure to an antigen, the animal body triggers an immune response and started to produce antibodies or proteins which only recognize and bind tightly to the specific antigens. Each antibody recognizes only a single antigen.
- Now, Scientists can produce antibodies by using the immune response of animals which can be used as tools for the detection and diagnosis of diseases.
- During the production of Primary Antibody, the antigens are first injected into an animal, after a period of time the serum is collected from the infected animal which will contain antibodies that specifically recognize that antigen.
- These antibodies can be used to develop diagnostic tests for the disease. In an immunoassay, the antibodies used to recognize antigens like disease agents are called primary antibodies.
Secondary Antibodies or Enzyme Labeled Antibodies
- In the dot blot or western blotting technique, the Secondary Antibodies recognize and bind to primary antibodies.
- Secondary antibodies are prepared in the same manner as primary antibodies and the antigen is antibodies from a different species, normally a fragment containing the constant (conserved) domain.
- A specific enzyme is chemically coupled to the constant domain of the antibody, which is away from the antigen binding domain. Mainly two types of enzymes are used for the coupling such as horseradish peroxidase (HRP) and alkaline phosphatase (AP).
- These enzymes catalyze a chemical substrate which leads to the formation of either chemiluminescence (light) or colorimetric (color) product that can be detected.
- This experiment uses HRP and a colorimetric substrate is known as 3,3’,5,5’‐ tetramethylbenzidine (TMB).
Material Required
- 30μl each of Simulated Sample 1, Simulated Sample 2, IMU Positive Control and IMU Negative Control.
- 1X MEM Washing Buffer (shared with class)
- 1X Blocking Buffer (NAP‐Blocker) (shared with class)
- 1 vial Antibody: BE Antibody 1
- 1 vial Antibody: BE Antibody 4 (HRP Secondary)
- 1 bottle of HRP Substrate (shared with class)
- 4 strips of Protein Binding Membrane
- 2 50mL tubes
- 1 12cm x 12cm washing tray
Dot Blot General Procedure
Spotting Sample Protocol
- Using forceps remove the nitrocellulose membrane strip from the protective cover. Place the strip on a clean flat surface (e.g. clean white paper). Label with your name at the left end of the strip with a pencil.
- Using a 5‐10μl pipette, remove 5μl of the sample and spot onto the membrane. With the tip end held 0.5cm/ ¼” above the area to be spotted, slowly push the sample out of the tip to form a hanging drop. Touch the drop onto the center of the area to be spotted and allow the sample to enter the membrane. Spot all the samples on the strip.
- Once all students in your group finished spotting and all liquid has absorbed onto the membrane, place all membrane strips in a 12x12cm washing tray with forceps.
Protein Detection
- Add 20ml 1X Blocking Buffer (NAP‐Blocker) to block the non‐specific sites. Incubate at room temperature for 30‐60 minutes with gentle shaking.
- Prepare the primary antibody by adding 40μl BE Antibody 1 to 20ml 1X Blocking Buffer (NAP‐Blocker).
- Discard the Blocking Buffer. Add the primary antibody solution to the membrane and incubate for 30‐60 minutes at room temperature with gentle shaking.
- Discard the antibody solution and wash 3 times with 20ml MEM Washing Buffer for 10 minutes each.
- Make secondary antibody by mixing 40μl BE Antibody 4 (HRP Secondary) with 20ml 1X Blocking Buffer (NAP‐Blocker) in a 50mL tube. The secondary antibody has a horseradish peroxidase tag.
- Discard the MEM Washing Buffer and add the secondary antibody solution to the membrane and incubate at room temperature for 30‐60 minutes with gentle shaking.
- Discard the antibody solution and wash 3 times with 20ml 1X MEM Washing Buffer for 10 minutes each.
- Discard the MEM Washing Buffer and add 5ml of HRP Substrate to the membrane. Let shake for 5 minutes or until color develops at room temperature.
- Pour off the substrate and add DI water to stop the color reaction. Record your results.
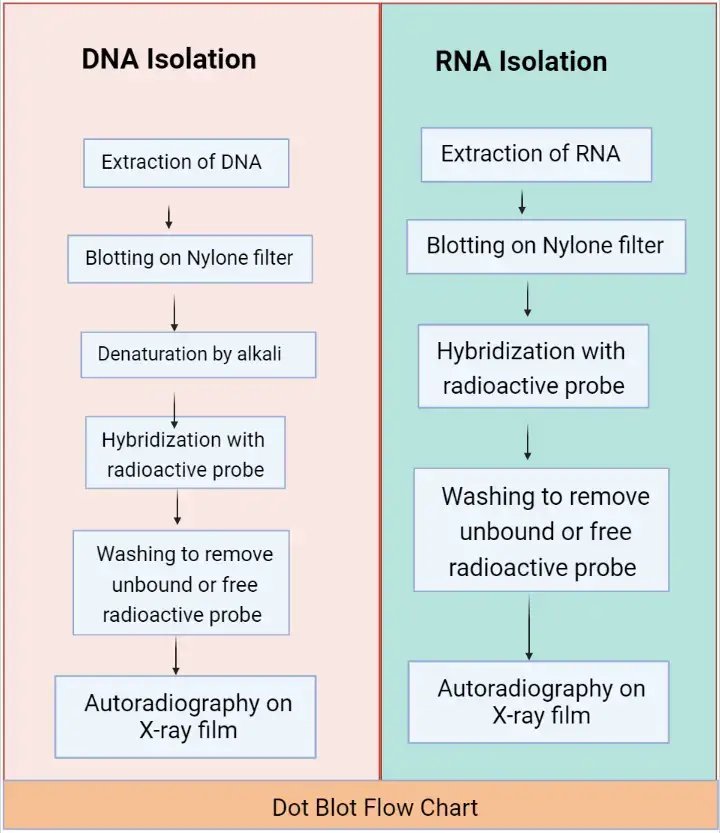
Dot Blot for RNA or DNA Isolation
The DNA identification by dot blot is performed by these following steps;
- Extraction of RNA/DNA: Collect samples of the RNA/DNA from the different tissues or cells.
- Blotting: Perform the blotting of collected RNA/DNA sample onto the nitrocellulose or nylon filter membrane.
- Denaturation (this step only for ds-DNA): The ds-DNA is passed through alkali treatment to convert it into single strands.
- Hybridization: Addition of a radioactive probe to the filter medium containing a DNA sample. This probe will bind to the mark DNA or later hybridize it.
- Washing: Wash the sample to remove the unbound or free radioactive probe from the filter medium.
- Autoradiography: Place the filter membrane to the X-ray film, after which one can visualize the aspired gene of DNA.
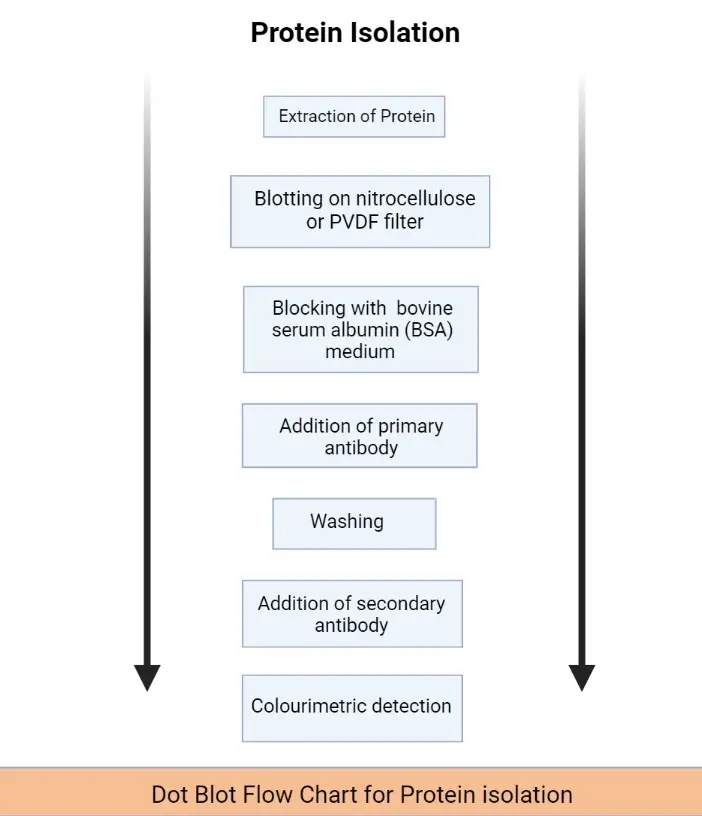
Dot Blot for Protein Isolation
- Extraction of Protein: Collect samples of the Protein sample from the different tissues or cells.
- Blotting: Add the collected protein sample onto the nitrocellulose or PVDF filter membrane.
- Blocking: Block the extracellular space in the filter membrane with the addition of bovine serum albumin (BSA) medium or dry milk.
- Addition of primary antibody: The primary antibodies fix with the target protein molecule.
- Washing: Wash the filter membrane to remove the unbound primary antibodies by using the PBS buffer.
- Addition of secondary antibody: This antibody only binds with the primary antibodies. The secondary antibodies are attached to the enzymes. After the attachment with the primary antibody, a substrate is added it will give a specific color to the sample.
- Colorimetric detection: Use the Colourimetric detection method to measure the intensity of color which is formed after the addition of substrate.
Dot blot Advantages
The dot blot technique is so similar to the western blotting technique but during the experiment we use prefers Western blotting instead of dot blot, because there are several drawbacks in western blotting such as;
- More materials: A dot blot requires all the same materials as western blotting needed but in a very low quantity such as a polyacrylamide gel, molecular weight ladder, loading dye, along with apparatuses and buffers.
- More steps: In the western blotting technique the sample needs to be load and run on a polyacrylamide gel and also need to transfer from gel to membrane, which is absent in the dot blot technique.
- More time-consuming process: The dot blot technique can be completed within 3 hours while western blotting requires two days to complete its al; steps including the transfer step and a possible overnight incubation with the antibody.
- Limited samples: The number of samples assessed in a Western is limited by the number of lanes in the polyacrylamide gel. A dot blot, by comparison, can screen a large number of samples for the presence of a POI at once.
Dot Blot Limitation
- The main drawback of dot blot is it cannot separate proteins by their size.
- This method does not typically use housekeeping proteins to normalize signal.
- As the protein in the sample has not been separated by size, this can make detection of a false positive (such as non-specific binding to peptide fragments) difficult to detect.
- A dot blot would not allow for comparison of a normal and modified target within the same blot.
References
- https://www.deshbandhucollege.ac.in/pdf/e-resources/botany/LS-VI-Blotting-Techniques.pdf
- https://en.wikipedia.org/wiki/Blot_(biology)
- https://www.rndsystems.com/resources/protocols/dot-blot-protocol
- https://www.abcam.com/ps/pdf/protocols/dot%20blot%20protocol.pdf
- https://www.slideshare.net/tailorparvez/blotting-techniques-95897728
- https://www.slideshare.net/TapeshwarYadav1/blotting-techniques1
- Principles of Dot Blots, Deric M. Griffin, Posted on Oct 31, 2017
- Supriya N. (2021, January 9). Dot Blot Technique – Definition, Process & Applications. Biology Reader. https://biologyreader.com/dot-blot-technique.html