What is Northern blotting?
Northern blotting is a technique in molecular biology that allows for the detection of specific RNA sequences in a complex mixture, offering valuable insights into gene expression patterns.
In 1977, James Alwine, David Kemp, and George Stark at Stanford University introduced a method that was cleverly named “Northern blot,” referencing the earlier “Southern blot” technique used for DNA detection.
The process consists of multiple essential stages:
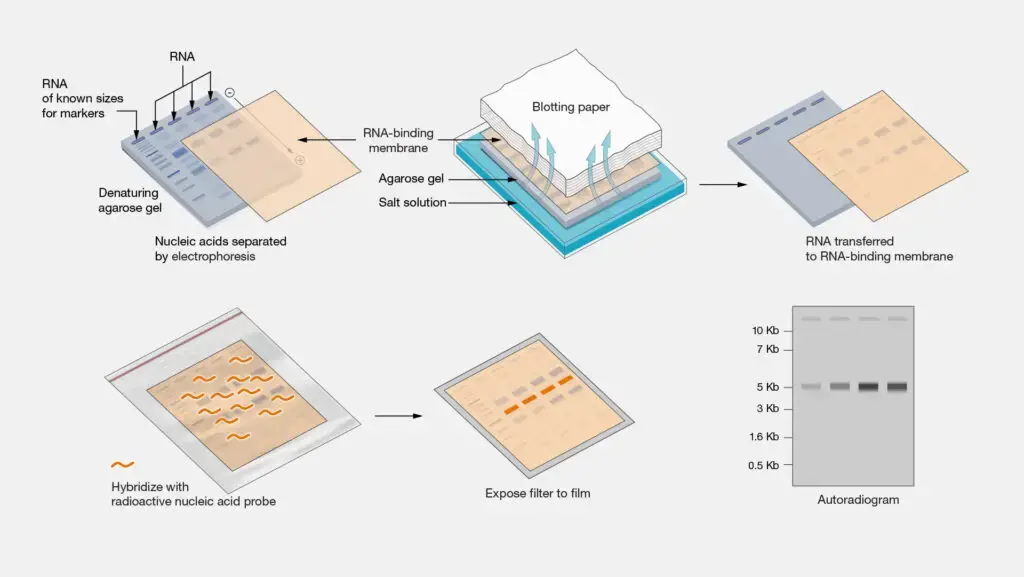
- Extraction of RNA from a biological specimen.
- RNA molecules are separated by size using denaturing gel electrophoresis, commonly employing formaldehyde-agarose gels to inhibit the formation of secondary structures.
- Transferring separated RNA onto a positively charged nylon or nitrocellulose membrane can be achieved through capillary action or vacuum transfer methods.
- Utilization of ultraviolet light or heat for the fixation of RNA to the membrane, ensuring stable binding.
- The membrane is hybridized with a labeled probe, which can be either radioactive or non-radioactive, designed to be complementary to the target RNA sequence.
- Utilization of autoradiography or chemiluminescence for the detection of the hybridized probe, facilitating the visualization of specific RNA bands.
Northern blotting plays a crucial role in:
- Evaluating gene expression levels in various tissues or during different developmental phases.
- Detecting alternative splicing occurrences through the analysis of RNA variants of varying sizes.
- Identifying RNA processing anomalies or alterations that influence transcript length.
- Confirming findings from alternative gene expression studies, including microarrays or RNA sequencing.
The benefits of Northern blotting encompass:
- Elevated specificity achieved through the application of sequence-specific probes.
- Capability to assess the dimensions of RNA transcripts, offering insights into transcript variants.
- Assessment of RNA integrity and quantity before proceeding with blotting.
- There is a possibility for membrane reprobing, enabling several analyses on a single sample.
Nonetheless, the method presents certain constraints:
- Exhibits reduced sensitivity in comparison to techniques such as RT-PCR, resulting in diminished effectiveness for identifying low-abundance transcripts.
- Procedures that require significant time and effort.
- There is a necessity for substantial quantities of high-quality RNA.
- There are potential hazards linked to radioactive labeling, despite the availability of non-radioactive alternatives.
Even with the development of more sophisticated methods, Northern blotting continues to be an important technique for examining gene expression, especially when details regarding transcript size and integrity are essential.
What is Southern blotting?
Southern blotting is a technique in molecular biology that was developed by Edwin Southern in 1975 to identify specific DNA sequences within a complex DNA sample.
The process consists of multiple essential stages:
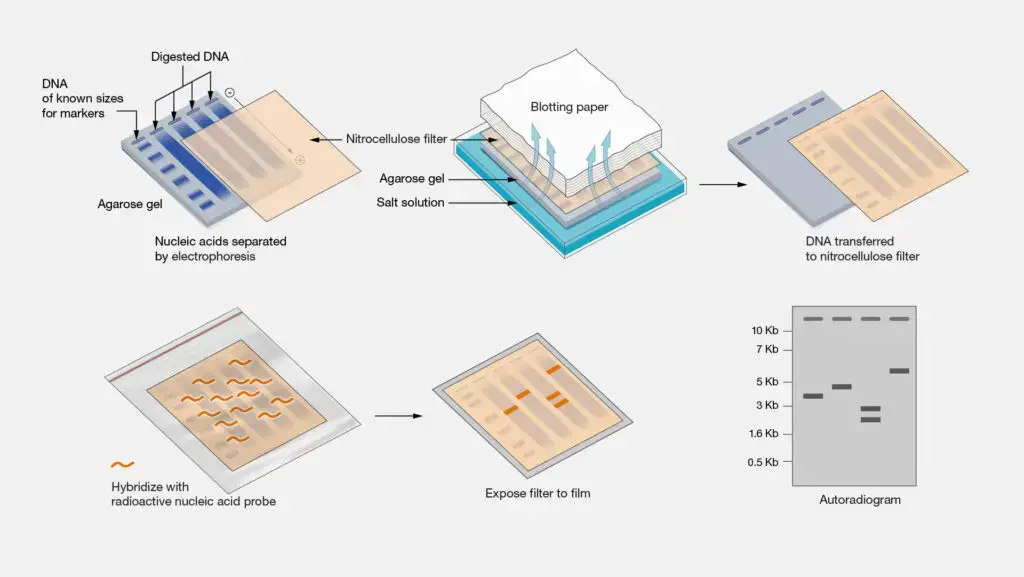
- Extraction of superior genomic DNA from the sample.
- Utilization of restriction enzymes for the digestion of DNA to produce fragments.
- Isolation of DNA segments through agarose gel electrophoresis.
- The process of denaturing DNA in the gel results in the formation of single-stranded DNA.
- Transferring DNA fragments onto a nitrocellulose or nylon membrane can be achieved through methods such as capillary action or vacuum transfer.
- Combining the membrane with a labeled DNA probe that is complementary to the target sequence.
- Identification of hybridized probes through autoradiography or chemiluminescence techniques.
Southern blotting plays a crucial role in:
- Mapping genes and identifying rearrangements within them.
- Identification of mutations, insertions, deletions, and structural variations.
- Examination of DNA methylation patterns.
- Identification of genetic conditions and infectious illnesses.
- Investigations in forensic science and testing for paternity.
The benefits of Southern blotting encompass:
- Elevated specificity achieved through the application of sequence-specific probes.
- Capability to assess the dimensions and number of DNA fragments.
- Identification of particular DNA sequences in intricate genomic landscapes.
Nonetheless, the method presents certain constraints:
- Procedures that require significant time and effort.
- Need for substantial quantities of high-quality DNA.
- Exhibits reduced sensitivity in comparison to PCR-based techniques.
- The utilization of radioactive materials raises significant safety issues; however, there are non-radioactive alternatives that can be considered.
Even with the advent of more sophisticated methods, Southern blotting continues to be an important technique for analyzing DNA, especially when details regarding fragment size and structural variations are essential.
Northern Blot vs Southern Blot
Here’s a detailed comparison between Northern blot and Southern blot –
| Feature | Northern Blot | Southern Blot |
|---|---|---|
| Target Molecule | RNA | DNA |
| Purpose | Detect and analyze gene expression levels | Identify and characterize specific DNA sequences |
| Sample Preparation | RNA extraction and purification required | DNA extraction and purification required |
| Denaturation | RNA is denatured with heat and/or formaldehyde | DNA is denatured with heat or alkali solution |
| Gel Electrophoresis | Agarose gel is used | Agarose or polyacrylamide gel is used |
| Transfer Method | Capillary or vacuum transfer | Capillary or vacuum transfer |
| Membrane Type | Nitrocellulose or nylon membrane | Nitrocellulose or nylon membrane |
| Probe Types | DNA, RNA, or oligodeoxynucleotides | Nucleic acid probe with a homologous sequence to the target molecule |
| Probe Labeling | Typically labeled with radioisotopes (e.g., 32P) or non-radioactive labels | Labeled with radioisotopes (e.g., 32P) or non-radioactive labels |
| Hybridization | RNA probe hybridizes with complementary RNA sequences | DNA probe hybridizes with complementary DNA sequences |
| Detection | Autoradiography or chemiluminescence | Autoradiography or chemiluminescence |
| Sensitivity | Less sensitive than Southern blotting | More sensitive than Northern blotting |
| Specificity | Detects specific RNA sequences | Detects specific DNA sequences |
| Applications | Gene expression analysis, RNA quantification | DNA fragment analysis, DNA mapping, genetic testing |
| Target Abundance | Can detect low-abundance RNA transcripts | Can detect low-abundance DNA sequences |
| RNA Stability | RNA is less stable and prone to degradation | DNA is more stable and less prone to degradation |
| Reverse Transcription | May involve reverse transcription step to convert RNA to DNA | Not applicable |
| Ribosomal RNA Removal | Usually requires removal of ribosomal RNA | Not necessary for most applications |
| Complexity | Generally less complex than Southern blotting | Generally more complex than Northern blotting |
| Availability | Less commonly used compared to Southern blotting | More commonly used compared to Northern blotting |
- https://www.aatbio.com/resources/faq-frequently-asked-questions/what-are-the-differences-between-northern-and-southern-blotting
- https://biologywise.com/northern-blot-vs-southern-blot-technique
- https://bio.libretexts.org/Bookshelves/Microbiology/Microbiology_%28Boundless%29/07%3A_Microbial_Genetics/7.25%3A_Molecular_Techniques/7.25C%3A_Northern_Blots
- https://www.enzo.com/note/what-are-the-differences-between-northern-southern-and-western-blotting/
- https://biologyease.com/types-of-blotting-techniques/
- https://www.sigmaaldrich.com/IN/en/technical-documents/protocol/protein-biology/gel-electrophoresis/southern-and-northern-blotting
- https://pediaa.com/difference-between-southern-northern-and-western-blotting/