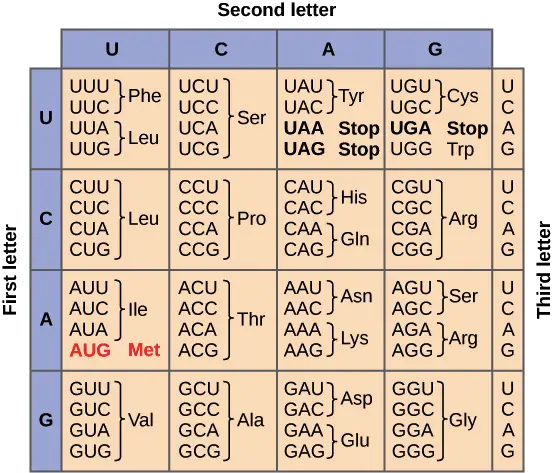
An codon chart is presented as a compact key, that maps three-nucleotide codons to amino acids, and it is usually shown as a table or circular map. In the chart, each codon is represented by three bases (A, U, G, C) and is read in the 5’→3’ direction, misreading the direction will alter meaning.
Sixty-four codons are listed, 61 are coding and 3 are stop codons (UAA, UAG, UGA), which are indicated to terminate translation. The feature of degeneracy is displayed where several codons specify the same amino acid, this redundancy is shown clearly on many charts.
During translation, codons are matched to amino acids by tRNA anticodons inside the ribosome, and the chart is used conceptually to know which residue is added.
Start and stop signals are highlighted (eg., AUG for start ; UAA/UAG/UGA for stop) so that the reading frame can be located quickly on lab sheets. Wobble at the third base is illustrated, which explains why some mutations at 3rd position are tolerated and protein change is sometimes avoided.
The code was deciphered in experiments with E. coli ( Escherichia coli ), and assignments were confirmed by later work, though exceptions were later noted. Charts are formatted as grids or circular diagrams, and they are annotated (colors, notes) so start/stop and redundant codons are easy to spot.
Reading frame is stressed, because a shifted frame produces nonsense or truncated proteins, it is a common source of error. Walking along the mRNA, the codons were skimmed quickly, the meaning sometimes got lost.
Practical use is shown in predicting protein sequence from mRNA and in designing site-directed mutagenesis / codon optimization for expression. The genetic code is regarded as largely universal, but variant codes are reported in mitochondria and some microbes, and these exceptions are shown on special charts.
The list of codons are printed on reference sheets, and spacing/inconsistent reading cause mistakes (dangling modifiers, run-ons happen).
Overall, a codon chart is provided as a concise translation map between nucleotide triplets and amino acids, simple in idea, but careful attention is required.
What is a Codon Chart?
- A codon chart is described as a key that translates three-nucleotide groups into amino acids, and it is presented usually as a table or circle.
- A codon is formed by three bases (triplet), and it is represented on the chart as combinations like AUG, GCU, UGA, etc.
- The chart is used to identify which amino acid will be added, when an mRNA codon is read by the ribosome, but sometimes readers get the frame wrong.
- mRNA codons are normally written in the 5’→3’ direction, though students sometimes read 5’ →3’ or 5’→ 3’ incorrectly, which changes meaning.
- Sixty four total codons are listed on the chart, 61 are coding and 3 are stop codons (UAA, UAG, UGA), and this was found experimentally.
- The concept of degeneracy (many codons → same amino acid) is shown on the chart, and it is relied upon to reduce error, yet confusion occurs.
- Translation is performed in the ribosome, and during translation, tRNA brings the appropriate amino acid — so the chart is consulted indirectly.
- The codon AUG is shown as start codon and codes for methionine (Met); it is also used to initiate translation.
- The code was deciphered by experiments (e.g., with _ _Escherichia coli ****) and by scientists like Nirenberg and Khorana, who mapped many codons.
- In most organisms the code is universal, but exceptions are noted in some mitochondria, and some microbes show variant codons.
- When a base substitution happens, a codon is changed, a different amino acid can be specified, and proteins may be altered, this is often checked against the chart.
- The chart is usually read first base = left, second = top, third = right (in circular or tabular forms), but misreading the order will give wrong results.
- The genetic code was found to be non-overlapping, meaning each base belongs to a single codon at a time, which prevents frame ambiguity.
- By using the codon chart, researchers can predict protein sequence from mRNA, and they do it routinely in labs / biotech.
- Such as, for codons that differ only at third base (wobble), redundancy is shown on chart, and it helps tolerate mutations.
- Walking along the mRNA, the codon was noted quickly by the student (dangling modifier), the meaning sometimes still missed.
- The list of codons are printed on lab sheets, and highlighting of start/stop is often done, to help visualize where translation begins / ends.
- It is emphasized that the reading frame must be correct, wrong frame = nonsense ; fragments, stop or shifted proteins will be produced.
- The chart may be circular or tabular, and they are used interchangeably by many, though some prefer tables for clarity.
- In short, a codon chart is provided as a map from nucleotide triplets to amino acids, it’s simple in concept, but mistakes are common if attention is not paid.
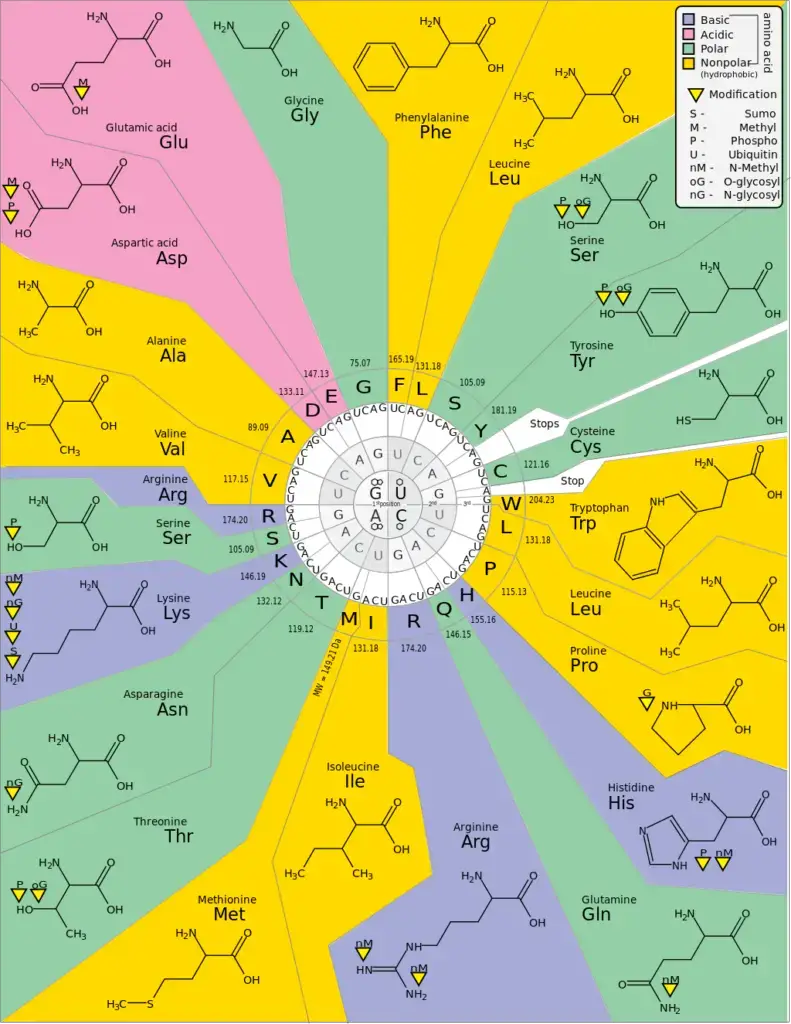
20 standard amino acids with their three-letter and one-letter abbreviations:
- Alanine (Ala, A)
- Arginine (Arg, R)
- Asparagine (Asn, N)
- Aspartic acid (Asp, D)
- Cysteine (Cys, C)
- Glutamic acid (Glu, E)
- Glutamine (Gln, Q)
- Glycine (Gly, G)
- Histidine (His, H)
- Isoleucine (Ile, I)
- Leucine (Leu, L)
- Lysine (Lys, K)
- Methionine (Met, M)
- Phenylalanine (Phe, F)
- Proline (Pro, P)
- Serine (Ser, S)
- Threonine (Thr, T)
- Tryptophan (Trp, W)
- Tyrosine (Tyr, Y)
- Valine (Val, V)
| Amino Acid | Abbreviation | One-Letter Abbreviation |
| Alanine | Ala | A |
| Arginine | Arg | R |
| Asparagine | Asn | N |
| Aspartic acid | Asp | D |
| Cysteine | Cys | C |
| Glutamic acid | Glu | E |
| Glutamine | Gln | Q |
| Glycine | Gly | G |
| Histidine | His | H |
| Isoleucine | Ile | I |
| Leucine | Leu | L |
| Lysine | Lys | K |
| Methionine | Met | M |
| Phenylalanine | Phe | F |
| Proline | Pro | P |
| Serine | Ser | S |
| Threonine | Thr | T |
| Tryptophan | Trp | W |
| Tyrosine | Tyr | Y |
| Valine | Val | V |
Codon Chart
| First Base | Second Base | U | C | A | G |
|---|---|---|---|---|---|
| U | U | UUU Phe | UCU Ser | UAU Tyr | UGU Cys |
| UUC | UCC | UAC | UGC | ||
| C | UUA Leu | UCA Ser | UAA STOP | UGA STOP | |
| UUG | UCG | UAG STOP | UGG Trp | ||
| C | U | CUU Leu | CCU Pro | CAU His | CGU Arg |
| CUC | CCC | CAC | CGC | ||
| C | CUA Leu | CCA Pro | CAA Gln | CGA Arg | |
| CUG | CCG | CAG | CGG | ||
| A | U | AUU Ile | ACU Thr | AAU Asn | AGU Ser |
| AUC | ACC | AAC | AGC | ||
| C | AUA Ile | ACA Thr | AAA Lys | AGA Arg | |
| AUG Met | ACG | AAG | AGG | ||
| G | U | GUU Val | GCU Ala | GAU Asp | GGU Gly |
| GUC | GCC | GAC | GGC | ||
| C | GUA Val | GCA Ala | GAA Glu | GGA Gly | |
| GUG | GCG | GAG | GGG |
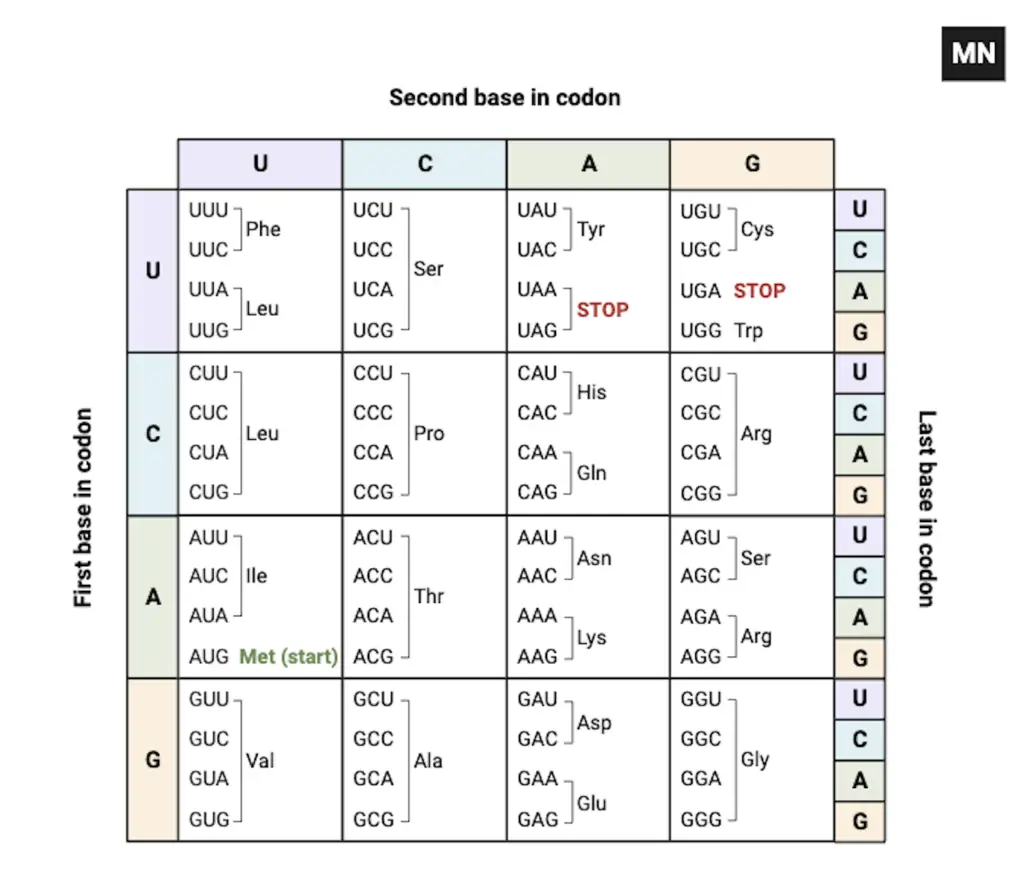
How do you read a codon table?
- Step 1: mRNA sequence should be located and oriented 5’→3’ on the page, because the codon table is read in that direction and misreading will change meaning.
- Step 2: The codon table (or chart) is to be consulted to match each three-base group, and the first base is taken from left, the second from top, the third from inner/right depending on chart layout.
- Step 3: AUG is identified as start codon.
- Step 4: The reading frame is established by choosing the correct start, otherwise a shifted frame will produce nonsense or truncated proteins.
- Step 5: Walking along the mRNA, the codon triplet is read sequentially and, by using the table, the corresponding amino acid is determined (eg., Met for AUG), the procedure is then repeated codon after codon.
- Step 6: Wobble at the third base is considered so that different codons are sometimes mapped to the same amino acid and degeneracy is noted on the chart.
- Step 7: Stop codons (UAA, UAG, UGA) are located as termination signals, and translation is halted when they are encountered.
- Step 8: For variant genetic codes, the mitochondrial or specialized tables are used, and E. coli (E. coli) was among the organisms used in early decoding experiments.
- Step 9: If a mutation is suspected, the altered base is compared with the table to see if a silent, missense, or nonsense change is produced, which helps infer functional impact.
- Step 10: The list of codons are printed on lab-sheets and emphasis is placed on correct spacing / reading, mistakes are common, such as, reading 3’ →5’ by accident.
- Step 11: A quick check is made by translating a short stretch (3–6 codons) to verify frame and expected residues before full translation is attempted.
- Step 12: Finally, the full protein sequence is assembled from the codon-derived residues and the output is verified against known sequences or predictions, done routinely in labs / bioinformatics.
Here is an example – How do you read a codon table?
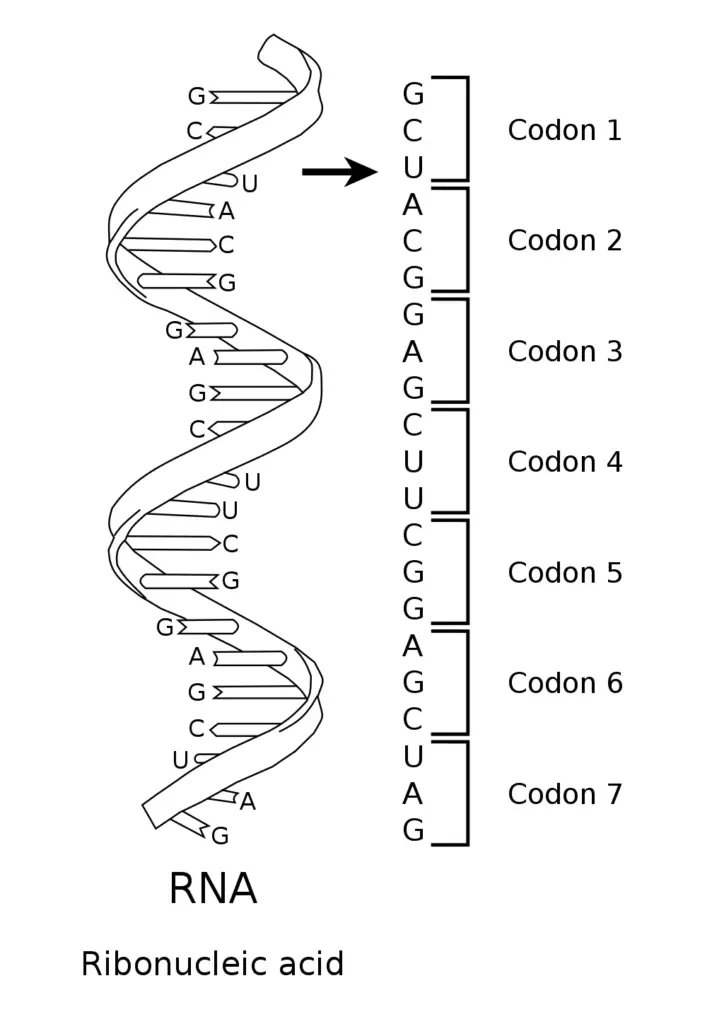
To read the codon chart, find the amino acid that forms a three-nucleotide sequence, or codon, in mRNA. The chart translates mRNA to its equivalent amino acid. Here’s how it works:
Here’s a clear step‑by‑step guide to reading the codon chart you provided:
- Start with an mRNA codon of 3 bases in 5′→3′ direction, e.g. “AGU”
- Find the first base on the left-hand axis (rows labeled U, C, A, G); for AGU you go to row A
- Then find the second base on the top axis (columns U, C, A, G); here it’s G, so locate intersection where row A meets column G
- Inside that grid box you’ll see 4 small entries, one for each possible third base; these are labeled U, C, A, G vertically or listed together
- Find your third base, here it’s U, so pick the entry “AGU”
- The entry tells you which amino acid or signal, e.g. “Ser” (serine)
- Note special cases:
- “AUG” is marked green as Met (start)
- “UAA, UAG, UGA” are red and labeled STOP
- Repeat for each consecutive triplet until a stop codon appears terminating translation
- If using a codon wheel, same logic applies: choose rings for first, second, third base from center
What are Start Codons?
- Start codons are defined as specific mRNA triplets that are recognized to begin translation, and they are usually indicated on sequences to set the reading frame.
- The most common start codon is AUG, which is shown to code for methionine (Met) in eukaryotes and for a formyl-methionine (fMet) in bacteria, but sometimes other codons are used.
- In bacteria (eg., E. coli (E. coli)) alternative starts like GUG or UUG are seen and these are used rarely, the ribosome may still initiate at them.
- The ribosome is guided to the start site by signals (such as, Kozak sequence in eukaryotes ; Shine-Dalgarno in prokaryotes) and it is then positioned so the codon is read in 5’ →3’ direction.
- tRNA carrying Met (or fMet) is recruited at initiation, and the first amino acid is thus brought, which anchors the polypeptide synthesis.
- Start codons are highlighted on gene maps and lab-sheets so translation start can be located quickly, errors in locating it lead to shifted frames and nonsense proteins.
- The reading frame is fixed by the start codon, wrong choice of start will produce entirely different amino acid sequence, and this is a frequent source of confusion.
- In some viruses and organelles variant genetic codes are used, therefore specialized tables are consulted and exceptions are noted.
- A short surrounding context is checked (eg., Kozak consensus: gccRccAUGG ), because context often determines which AUG is used, even if several AUGs are present.
- Initiation factors (IFs) are involved in the process, they assist ribosome assembly and start site selection, but the factors vary between bacteria and eukaryotes.
- When mutation alters the start codon, initiation may be abolished or shifted, leading to truncated proteins or to use of downstream starts, which can be deleterious.
- Practically, the start codon is read first when translating an ORF (open reading frame), a quick check of first 3 bases is done to confirm, then translation is continued codon-by-codon.
- In summary, a start codon is seen as the signal that tells the cell where to begin translating mRNA into protein, it is small but crucial.
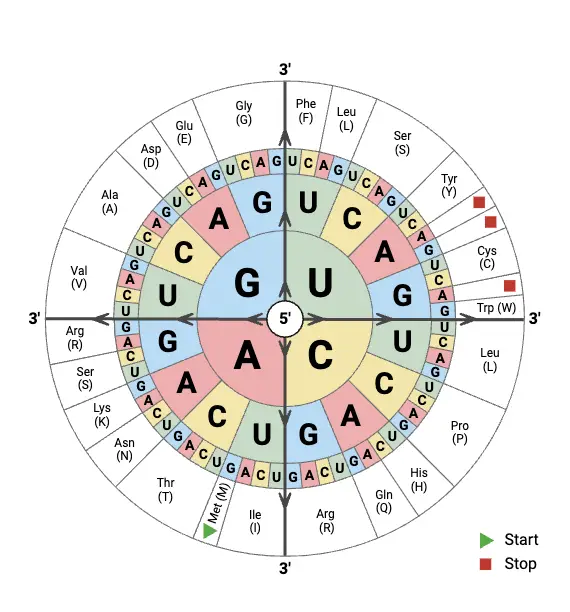
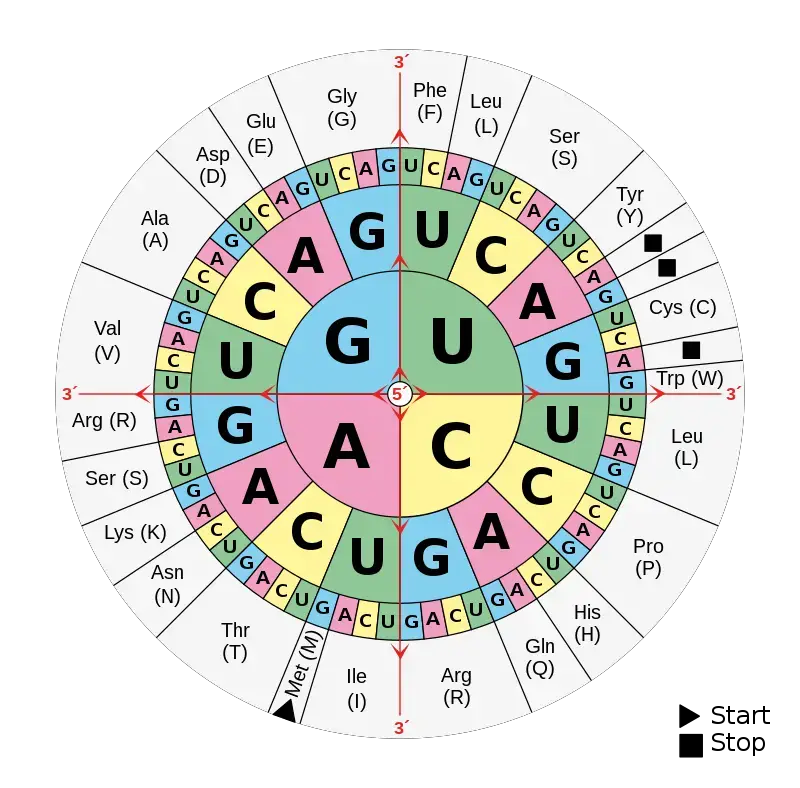
How to Read a Codon Wheel?
- A codon wheel is used to determine which amino acid is specified by an mRNA codon, and it is read from the center outward, not the other way round.
- The first base of the codon (5’ position) is found in the innermost circle of the wheel, and this base directs the starting section to be followed.
- From that starting base, the second base is traced outward to the middle ring, which narrows the options toward a smaller section of the wheel.
- The third base, which is located on the outermost ring, is then chosen and it finally points to a single amino acid or a stop signal.
- Each codon (3-letter combination) is therefore traced as a path from center → middle → outer edge, and at the end of the path the corresponding amino acid name/symbol is shown.
- For example, to read AUG: start at inner “A,” then go to “U” in middle, then “G” on outer ring, it leads to Methionine (Met) which also acts as the Start Codon.
- Sometimes mistakes are made because the wheel is read outside-in, or codons are written from the wrong strand (DNA instead of mRNA), causing mismatch.
- Stop codons like UAA, UAG, UGA are marked on the wheel (often as STOP or with a star *) and these are identified as termination signals during translation.
- When reading the wheel, codons are always interpreted in the 5’→3’ direction, though beginners sometimes reverse it or mix with tRNA anticodons.
- Redundancy is shown clearly because several outer segments (different codons) point to the same amino acid, illustrating degeneracy of the code.
- In many charts, color shading / bold outlines are used for grouping codons coding for same amino acids, which makes visual recognition quicker.
- The wheel is designed to make it easy to find amino acids for any of the 64 codons, but it requires slow reading at first until the direction becomes familiar.
- In practice, codon wheels are printed or drawn on lab-notebooks, and they are referenced during gene translation, mutation study or synthetic biology design.
- Walking through the wheel from center to edge, the codon sequence is literally built in reading order, though sometimes the eyes skip lines, producing errors.
- In summary, to read a codon wheel, one should start at inner circle (1st base), move to middle (2nd), finish at outer (3rd) and then read the amino acid / stop that’s aligned on that path — simple but needs care.
What are Stop Codons?
- Stop codons are triplet nucleotide sequences in mRNA (messenger RNA), which are defined to signal the termination of translation / protein synthesis.
- The three main stop codons — UAA, UAG, and UGA — are typically present in most organisms, and are recognized by release factors, not by tRNAs, this distinction is important.
- Released at the ribosome, proteins are freed when a stop codon is encountered, leading to translation termination, a process that is mediated by factors and hydrolysis of the peptidyl-tRNA bond (slightly technical, yes).
- Release factors (RFs) are the molecules that recognize them, they promote chain release.
- In bacteria recognition is done by RF1 or RF2 depending on codon, while in eukaryotes a single eRF1 is involved, and context (neighboring bases) influences efficiency.
- The list of codons are described as simple, yet their biological consequence is large and sometimes, surprising (readthrough occurs).
- Though considered signals for stopping, they are sometimes bypassed by readthrough (when near-cognate tRNAs insert an amino acid), and this mechanism is exploited by some viruses / genes to produce extended proteins.
- No amino acid is encoded by a stop codon, so translation is terminated (short, clear).
- In model organisms like Escherichia coli (E. coli) and Saccharomyces cerevisiae (S. cerevisiae), stop codons are conserved, but the efficiency, context-dependence and how they are interpreted by the ribosome it/they varies.
What is Codon Translation?
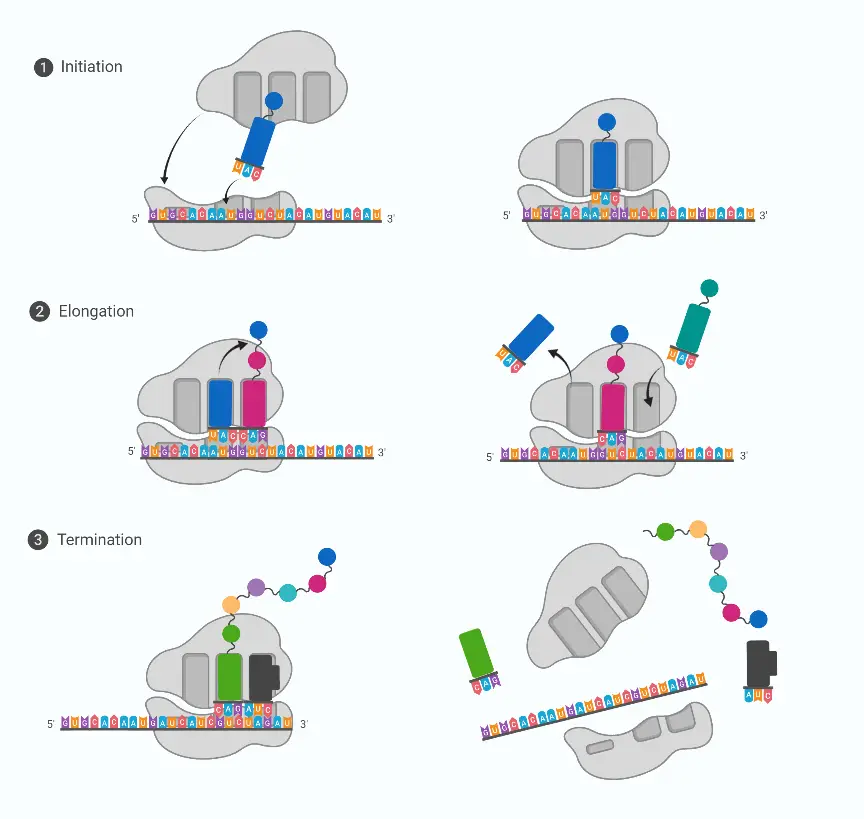
Codon Translation is the process by which mRNA triplets are read, and are converted into a chain of amino acids.
In this process, the sequence of codons (3 bases each) is usually read by the ribosome, and matching is performed by tRNA anticodons, though context affects accuracy.
Initiation is begun when a start codon (often AUG) is recognized, and methionine is placed as first residue (in bacteria fMet may be used).
Elongation is carried out as tRNAs are brought to the A-site, peptide bonds are formed by peptidyl transferase, and the chain is transferred to next tRNA, simple in idea but messy in practice.
Termination is signaled when a stop codon is encountered, and release factors bind causing release of polypeptide, no amino acid is added at that moment.
The ribosome moves along mRNA in 5’→3’ direction, pauses can occur, they influence folding and expression levels, and errors sometimes slip through, causing misincorporation.
The genetic code is mostly universal, though exceptions are present in mitochondria and some microbes, such as UGA coding Trp in human mito, small but important.
In Escherichia coli (E. coli) and Saccharomyces cerevisiae (S. cerevisiae) the translation machinery is conserved, yet regulation and factors differ between prokaryotes and eukaryotes.
In eukaryotes translation is separated from transcription (nucleus vs cytoplasm), while in prokaryotes they are often simultaneous, which changes control strategies.
Energy in the form of GTP is consumed during elongation and translocation, and initiation factors (IFs), elongation factors (EFs) and release factors are required, though names vary by species.
Ribosomes read codons, and they add amino acids brought by tRNAs, sometimes frameshifts happen, mistakes occur, causing truncated or faulty proteins.
Readthrough/recoding is sometimes exploited (by viruses, or regulatory genes) where a stop codon is bypassed, resulting in extended proteins; this is used biologically.
Errors are corrected to some extent by proofreading and quality-control systems, but complete fidelity is not achieved, so misfolding can follow.
Walking along mRNA, mistakes were made by the translational apparatus in some cases, a dangling thought but it reflects real occasional errors.
The process links DNA-encoded information to functional proteins, so genotype is translated into phenotype, simple bridge, crucial for life.
What is dna codon chart?
- A DNA codon chart is a table that is used to show which three-base codon (triplet of nucleotides) corresponds to an amino acid, and it is presented so that translation can be inferred from DNA sequence.
- Unlike the mRNA chart that uses U (uracil), a DNA chart uses T (thymine) in place of U, so codons are written with A, T, C, G instead of A, U, C, G.
- The triplets are read in a specific reading frame, and if shifted the meaning will be changed, causing different amino acids to be encoded; this is why frameshifts are critical.
- Start codon is shown as ATG (codes for Methionine, Met) and the stop codons are listed as TAA, TAG, TGA, these terminate translation.
- It is used to predict the peptide that will be produced after transcription/translation, though actual translation is performed on mRNA (messenger RNA) after transcription, a slightly indirect step.
- Redundancy (degeneracy) of code is indicated in the chart where multiple codons are mapped to same amino acid, so the genetic code is said to be degenerate.
- Universality is suggested by the chart, but exceptions are noted in mitochondria and some microbes, for example in human mitochondria they differ for a few codons, and in Escherichia coli (E. coli) small variations are sometimes observed.
- A simple 4x4x4 layout or a rectangular table is commonly used to represent it, and often codons are grouped by first/second/third base positions, which makes lookup easier.
- Walking along a sequence, a stop codon was noticed by the reader, dangling in the middle, an awkward note but it happens.
- Helpful in genetics, cloning, and in-silico translation / annotation; the chart is consulted by students, researchers, and they use it for designing primers / predicting proteins.
What is mRNA codon chart?
A mRNA codon chart is a table that is used to map each three-base codon in mRNA (messenger RNA) to its corresponding amino acid, and it is consulted during translation.
In the chart, codons are written with U (uracil) rather than T, and the start codon (AUG) plus stop codons (UAA, UAG, UGA) are usually highlighted, which helps to find ORFs.
Reading frames are indicated because when shifted the meaning will change, causing frameshifts and different polypeptides to be predicted, so care must be taken.
The genetic code is shown as largely universal, though exceptions (eg., mitochondria) are noted; such as, UGA → Trp in human mito, and this is sometimes pointed out.
A simple grid or 4×4×4 layout is commonly used, and the list of codons are arranged by first/second/third base, making lookup easier though the formatting is not always neat.
For practical work, it is used by students / researchers to translate sequences in-silico, and primers / constructs are designed with its aid.
Degeneracy of the code is shown where several codons map to the same amino acid, so redundancy is made explicit on the chart.
In model organisms like Escherichia coli (E. coli) and Saccharomyces cerevisiae (S. cerevisiae) the chart is applied, but codon usage bias and regulation differ between species.
Walking along an mRNA, the chart was glanced at, a dangling note but it reflects how annotation is sometimes done in quick lab notes.
It is noted that neighboring bases, codon usage, and mRNA secondary-structure influence translation efficiency, and sometimes recoding/readthrough events are predicted from context.
They are essential for teaching, annotation, cloning; simple, yet powerful.
Amino acid Codon Related Online Tools
- Amino Acid to Codon Mapper
- Codon Wheel Explorer
- Codon to Amino Acid Translator
- Interactive Codon Chart Tool
- Aseem, A. (2025, April 5). Codon Chart and Codon Wheel – explained. BiotechReality – Covering Industry, Business, and Academia. https://www.biotechreality.com/2024/11/codon-chart-and-codon-wheel-explained.html
- Codon Usage Frequency Table(chart)-Genscript. (n.d.). https://www.genscript.com/tools/codon-frequency-table
- Codon Table. (n.d.). https://www.genscript.com/tools/codon-table
- Indiana Biology Standards Resource. (2003). MRNA Codon/Amino Acid Chart. In B.1.23, B.1.26 / Curriculum Framework / How Do Mutations Affect Living Organisms?, How Does Protein Synthesis Occur? Indiana Biology Standards Resource. https://www.shsu.edu/academics/agricultural-sciences-and-engineering-technology/documents/mRNAcodonchart.pdf
- Genetic code & how to read a CoDOn chart – lesson | Study.com. (n.d.). study.com. https://study.com/academy/lesson/making-sense-of-the-genetic-code-codon-recognition.html
- Tamang, S. (2024, December 25). Codon Chart: Table, Amino Acids & RNA Wheel Explained. Microbe Notes. https://microbenotes.com/codon-chart-table-amino-acids/
- Codon-Amino acid abbreviations. (n.d.). https://www.hgmd.cf.ac.uk/docs/cd_amino.html
- Codon Table. (n.d.). SlideShare. https://www.slideshare.net/slideshow/howtoreadacodontable/40889076
- The Editors of Encyclopaedia Britannica. (2025, July 12). Genetic code | Definition, Characteristics, Table, & Facts. Encyclopedia Britannica. https://www.britannica.com/science/genetic-code