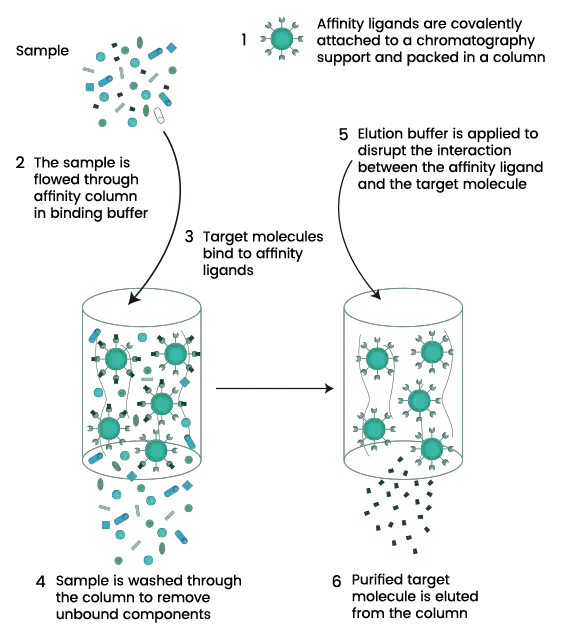
Affinity chromatography is a type of liquid chromatography technique which is used for separation and purification of biomolecules based on a highly specific and reversible interaction between the target molecule and a specific ligand. It is the process where the target molecule selectively binds with a ligand that is immobilized on a solid matrix.
This interaction is often compared with lock and key mechanism where ligand acts as lock and the specific binding site of target molecule acts as key. It is different from other chromatographic techniques because separation is done on the basis of biological activity or specific chemical structure and not on size or charge.
In affinity chromatography, the sample containing mixture of biomolecules is passed through a column packed with affinity matrix. Under suitable conditions, the desired molecule binds specifically to the ligand while other unbound components is washed out. After washing step, the bound target molecule is recovered by elution. Elution is carried out by changing the buffer conditions such as pH ionic strength or polarity or by adding a competitive ligand. This causes disruption of ligand–target interaction and the purified molecule is released.
An affinity chromatography system mainly consists of three components namely matrix ligand and spacer arm. The matrix is an inert solid support such as agarose or synthetic polymer which holds the ligand. The ligand is the molecule that specifically binds with target such as enzyme antibody or metal ion. The spacer arm is used to attach ligand to the matrix and it helps in reducing steric hindrance so that binding site is easily accessible.
Affinity chromatography is widely used for purification of specific proteins like antibodies enzymes and recombinant proteins. It is also used for removal of specific contaminants from biological samples. Some common types are Immobilized Metal Ion Affinity Chromatography (IMAC) and immunoaffinity chromatography. Due to its high specificity, this technique can provide very high degree of purification in a single step.
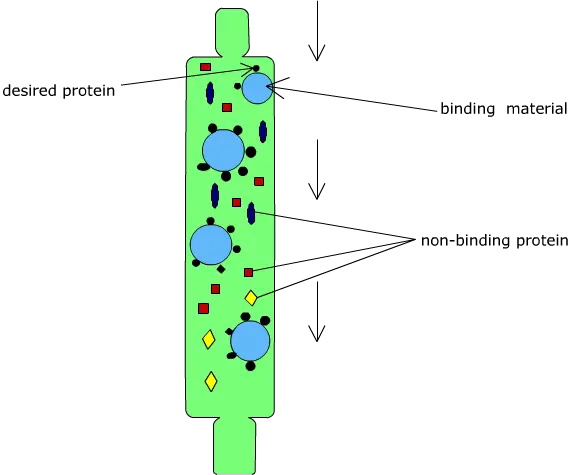
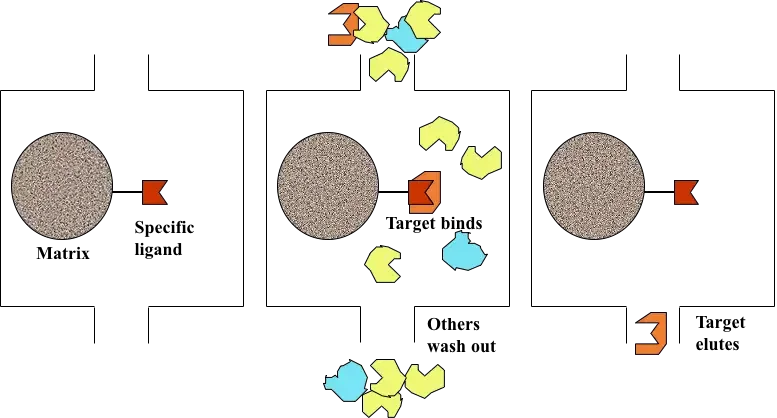
Principle of Affinity chromatography
Affinity chromatography is based on the principle of a highly specific and reversible biological interaction between a target molecule present in the sample and a specific ligand which is attached to a solid support. It is a liquid chromatography technique in which separation is carried out on the basis of biological function or individual chemical structure and not on physical properties like size or charge. In this method, the ligand present on the stationary phase selectively binds the desired molecule from the mobile phase. This interaction is commonly explained by lock and key mechanism where ligand acts as lock and the target molecule acts as key.
The binding between the ligand and the target molecule occurs due to non-covalent interactions such as hydrogen bonding electrostatic attraction and van der Waals forces. These interactions are specific and strong enough to retain the required molecule on the column but are reversible in nature. When the sample is applied under suitable buffer conditions, the target molecule binds to the ligand while the remaining components of the mixture does not show affinity and passes through the column.
After binding, the unbound substances are removed by washing and the bound molecule is then recovered by elution. Elution is achieved by altering the conditions that disturb the ligand-target interaction. It can be done by changing pH ionic strength or polarity of the buffer or by adding a competitive ligand. Thus the principle of affinity chromatography depends upon specific biological recognition between ligand and target molecule which allows high degree of purification in a single step.
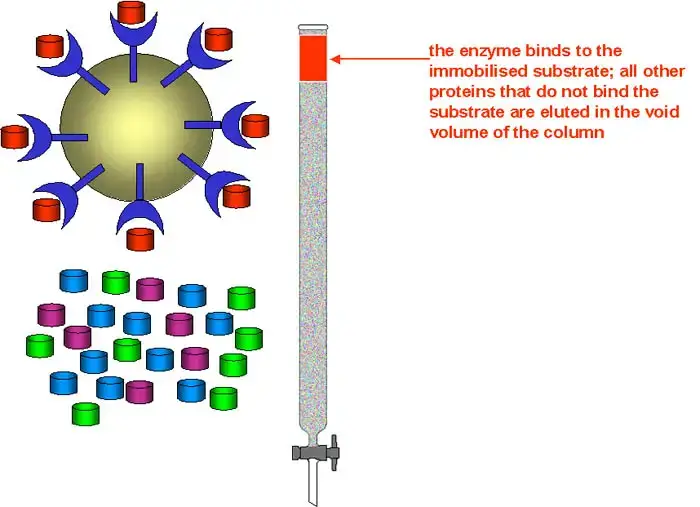
Components of affinity chromatography
The different components of affinity chromatography are described below–
1. Matrix (Solid support)– The matrix is an inert solid support to which the ligand is attached. It provides a large surface area for binding of target molecules and allows easy flow of the mobile phase. The matrix should be chemically stable and should not show non-specific adsorption of proteins. Commonly used matrices are agarose dextran cellulose and synthetic polymers.
2. Ligand– The ligand is the specific molecule which is covalently attached to the matrix and binds selectively with the target molecule. It acts as lock in the lock and key mechanism. The ligand may be an antibody enzyme receptor dye or metal ion depending upon the type of molecule to be purified. The interaction between ligand and target is specific and reversible.
3. Spacer arm– The spacer arm is a chemical chain that connects the ligand to the matrix. It helps in keeping the ligand away from the surface of matrix and reduces steric hindrance. This ensures that the binding site of ligand is easily accessible to the target molecule. In some cases absence of spacer arm may reduce binding efficiency.
4. Mobile phase (Buffer system)– The mobile phase consists of different buffer solutions used during the process. The binding buffer provides suitable conditions for interaction between ligand and target molecule. The wash buffer helps in removing unbound impurities. The elution buffer is used to break the ligand–target interaction by changing pH ionic strength or by adding a competitive agent.
5. Column– The column is the container in which the matrix with attached ligand is packed. It supports the stationary phase and allows controlled flow of the sample and buffers. In some methods magnetic beads are used instead of packed columns.
6. Target molecule– The target molecule is the desired biomolecule present in the sample which shows specific affinity towards the ligand. It may be a protein antibody enzyme or nucleic acid which is selectively retained and later purified.
Types of Affinity Chromatography
Different types of affinity chromatography are classified on the basis of nature of ligand and type of interaction involved. These are as follows–
A. Bioaffinity chromatography (Biospecific adsorption)
It is the type of affinity chromatography which uses naturally occurring biological molecules as ligands. These ligands show specific interaction with the target molecule and allow selective separation. This type includes antigen–antibody enzyme–substrate and receptor–ligand interactions.
- Immunoaffinity chromatography– It is based on specific binding between antigen and antibody. Either antigen or antibody is immobilized on the matrix. It is commonly used for purification of antigens antibodies and antibody–binding proteins such as Protein A and Protein G.
- Lectin affinity chromatography– This method uses lectins which bind specifically to carbohydrate moieties. It is used for separation of glycoproteins polysaccharides and glycolipids. Concanavalin A and wheat germ lectin are commonly used ligands.
- DNA or nucleic acid affinity chromatography– In this type DNA or RNA sequences are used as ligands. It is applied for purification of DNA-binding proteins such as polymerases transcription factors and restriction enzymes.
- Avidin–biotin affinity chromatography– It is based on strong interaction between avidin or streptavidin and biotin. This interaction is highly specific and stable and is used for purification of biotinylated proteins enzymes and nucleic acids.
B. Immobilized metal ion affinity chromatography (IMAC)
This technique is based on affinity of certain amino acid residues such as histidine cysteine and tryptophan towards metal ions. Metal ions like Ni²⁺ Cu²⁺ and Zn²⁺ are immobilized on the matrix. It is widely used for purification of His-tagged recombinant proteins.
C. Recombinant fusion protein affinity chromatography
This type is used for purification of proteins fused with specific affinity tags.
- Glutathione affinity chromatography– It is used for purification of GST-tagged proteins. Glutathione is immobilized on the matrix and binds specifically to glutathione-S-transferase.
- Maltose binding protein affinity chromatography– In this method amylose is used as ligand to purify proteins fused with maltose binding protein tag.
- Epitope tag affinity chromatography– It uses immobilized antibodies to bind short peptide tags such as FLAG Myc or HA fused to target proteins.
D. Pseudo-affinity chromatography
This method uses synthetic ligands which mimic natural biological substrates. These ligands show affinity towards a group of biomolecules rather than one specific molecule.
- Dye-ligand affinity chromatography– It uses synthetic dyes such as Cibacron blue which mimic enzyme cofactors. It is commonly used for purification of enzymes and albumin.
- Heparin affinity chromatography– Heparin is used as ligand which binds to DNA-binding proteins lipoproteins and coagulation factors.
E. Covalent affinity chromatography
This method is based on formation of reversible covalent bond between target molecule and matrix. It is mainly used for separation of thiol-containing compounds.
F. Boronate affinity chromatography
It is used for purification of molecules containing cis-diol groups such as carbohydrates nucleotides and glycoproteins. Boronic acid derivatives are used as ligands.
G. Cell affinity chromatography
This type is used for separation of whole cells based on specific receptors present on cell surface. It is mainly applied in immunological studies.
H. Molecularly imprinted polymer affinity chromatography
In this method synthetic polymers with specific cavities are used as artificial ligands. These cavities are complementary to the target molecule in shape and chemical nature.
How to prepare the Sample for Affinity Chromatography?
The sample must be properly prepared before loading on the affinity column. The main steps involved in sample preparation are as follows–
- Clarification of sample– The crude sample is first clarified to remove cells cell debris and insoluble materials. It is usually done by centrifugation so that suspended particles are removed. This step is important to avoid clogging of the column.
- Filtration– After centrifugation the sample is filtered to remove fine particulate matter. Filtration gives a clear solution and protects the affinity matrix from physical damage. It also helps in maintaining uniform flow through the column.
- Adjustment of buffer conditions– The sample is adjusted to the pH and ionic strength of the binding buffer. This is necessary for proper interaction between the ligand and the target molecule. Adjustment can be done by dilution with binding buffer or by buffer exchange.
- Solubilization of sample– The sample should be completely soluble before application. If the target molecule is insoluble suitable solubilizing agents may be used depending on the stability of the affinity matrix.
- Removal of interfering substances– Substances that interfere with binding or degrade the target molecule should be removed. Proteases lipids and other inhibitory compounds are eliminated as they reduce binding efficiency and affect purification.
- Desalting and buffer exchange– If the sample contains high salt concentration or small molecular weight impurities desalting is carried out. This step helps in transferring the sample into suitable binding buffer conditions.
- Concentration of sample (if required)– In some cases the sample is concentrated before loading. Concentration helps in reducing sample volume and increasing the amount of target molecule. This step is optional and depends on nature of sample.
Procedure of Affinity Chromatography (affinity chromatography steps)
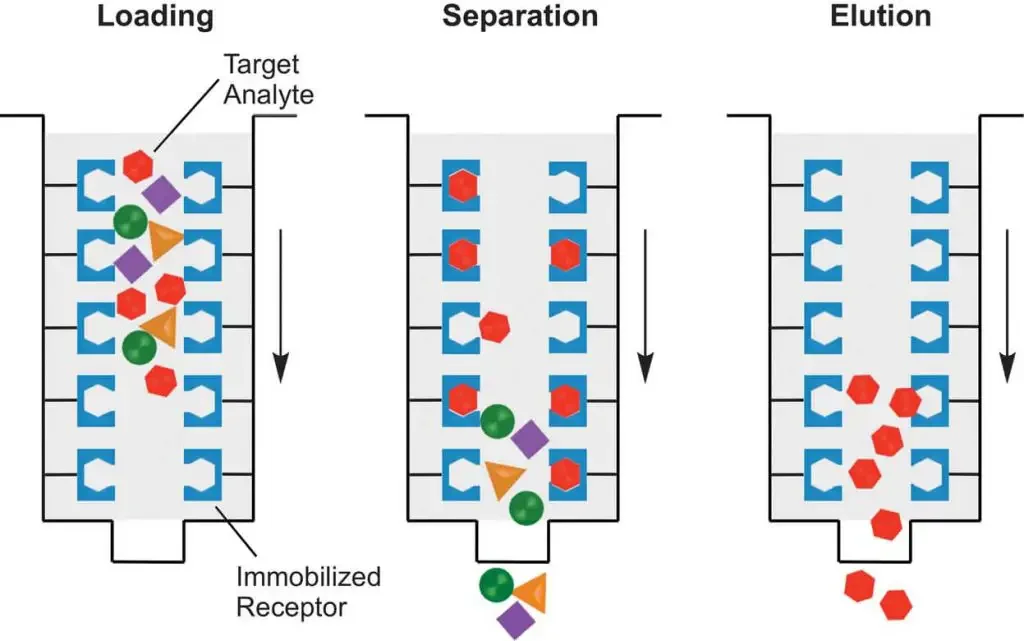
The procedure of affinity chromatography is carried out in a sequential manner and the steps are as follows–
- Column preparation and equilibration– The affinity matrix containing specific ligand is packed into the chromatography column. The storage solution present in the matrix is washed out properly using distilled water or suitable buffer. After this the column is equilibrated with binding buffer. This step is required to maintain proper pH and ionic strength for specific binding.
- Sample preparation– The crude sample such as cell lysate is clarified by centrifugation or filtration to remove insoluble particles. The pH and salt concentration of the sample is adjusted similar to binding buffer so that efficient binding can occur.
- Sample application (binding step)– The prepared sample is slowly applied to the equilibrated column. Under suitable conditions the target molecule binds specifically with the immobilized ligand present on the matrix. Other molecules which do not have affinity passes through the column.
- Washing of column– The column is washed with binding buffer to remove unbound and weakly bound impurities. Washing is continued until no unwanted protein is present in the eluent. This step helps in increasing the purity of bound molecule.
- Elution of bound molecule– The bound target molecule is eluted by changing the buffer conditions. This can be done by altering pH ionic strength or by adding a competitive ligand. The interaction between ligand and target molecule is disrupted and the desired molecule is released from the column.
- Collection of fractions– The eluted fractions containing purified target molecule is collected separately. If required neutralizing buffer is used to prevent denaturation of protein during elution.
- Column regeneration and re-equilibration– After elution the column is washed with suitable regenerating solution to remove tightly bound contaminants. The column is again equilibrated with binding buffer and can be reused for next cycle.
Elution Methods of Affinity Chromatography and Its Steps
Elution is the process by which the bound target molecule is released from the affinity matrix by disturbing the interaction between ligand and target. The elution methods used in affinity chromatography and their steps are described below–
Elution Methods of Affinity Chromatography
- Non-specific (non-selective) elution– In this method the physical or chemical conditions of the buffer are changed to weaken the interaction between ligand and target molecule.
Commonly used non-specific elution methods are–- Change in pH of bufferIncrease in ionic strengthUse of chaotropic agentsReduction of buffer polarity
- Specific (biospecific or competitive) elution– This method involves addition of a competitive molecule that has affinity for ligand or target. The competitive ligand replaces the bound target molecule and releases it from the column.
This method is more selective and usually gives higher purity and better activity of protein. - Step elution– in step elution the elution buffer is applied suddenly at a fixed concentration or pH. The bound protein is released in a short time and collected in concentrated form. This method is suitable for routine purification.
- Gradient elution– In gradient elution the concentration of eluting agent or pH is changed gradually. This provides better separation of closely related molecules and gives higher resolution.
Steps of Elution in Affinity Chromatography
- Selection of elution method– The appropriate elution method is selected based on strength and nature of ligand-target interaction. Mild conditions are preferred to preserve protein activity.
- Preparation of elution buffer– Elution buffer is prepared with required pH salt concentration or competitive ligand. The buffer should be freshly prepared and filtered.
- Application of elution buffer– The elution buffer is passed through the column after washing step. The change in conditions weakens the binding between ligand and target molecule.
- Release of bound target molecule– Due to altered conditions or competitive binding the target molecule dissociates from ligand and moves out of the column with eluent.
- Collection of eluted fractions– The eluted protein is collected in separate fractions. In case of low pH elution immediate neutralization is required to prevent denaturation.
- Post-elution washing and regeneration– After elution the column is washed thoroughly and regenerated using suitable buffer so that it can be reused for further purification cycles.
Factors Affecting Affinity Chromatography
The efficiency of affinity chromatography depends on several factors which directly influence binding specificity purification and recovery. The important factors are listed below–
- Nature of ligand– The ligand must be highly specific to the target molecule. If specificity is low non-specific binding occurs and purity is reduced. The strength of interaction between ligand and target is also important. Very weak interaction results in loss of target during washing while very strong interaction makes elution difficult. Ligand concentration on matrix also affects binding as very high density may cause steric hindrance.
- Matrix or solid support– The matrix used should be chemically inert so that non-specific adsorption of proteins does not occur. It must have proper porosity so that large biomolecules can easily enter and interact with ligand. The stability of matrix is also important to tolerate changes in pH ionic strength and flow rate during elution and regeneration.
- Spacer arm– In many cases a spacer arm is used to attach ligand to the matrix. If the spacer arm is too short the binding site of ligand may not be accessible to target molecule. If it is too long it may lead to non-specific interactions. The chemical nature of spacer arm also affects adsorption behaviour.
- pH and ionic strength of buffer– The binding between ligand and target is strongly influenced by pH and salt concentration. Proper pH is required to maintain native structure of protein and effective interaction. High ionic strength may weaken electrostatic interactions while very low ionic strength may increase non-specific binding.
- Sample composition– The sample applied to column should be free from particulate matter. Presence of lipids or high amount of impurities may block the column and interfere with binding. The pH and salt concentration of sample should match the binding buffer.
- Flow rate – Flow rate determines the contact time between ligand and target molecule. Very high flow rate reduces binding efficiency as sufficient interaction time is not provided. Lower flow rate improves binding especially for weak affinity interactions.
- Elution conditions– Elution of bound molecule depends on change in pH ionic strength or addition of competitive ligand. Harsh elution conditions may denature protein and reduce activity. Proper elution strategy helps in achieving high recovery and purity.
- Ligand immobilization method– The method of coupling ligand to matrix affects its orientation. Improper orientation may block the active binding site of ligand. Stable coupling is required to prevent ligand leakage into the final purified product.
Types of affinity media used in Affinity Chromatography
Affinity chromatography uses specific media for separation of molecules. These media is based on ligands attached to matrix. It is classified by ligand type and support material.
Types by Ligand Specificity
A. Bioaffinity media utilizes biological interactions.
These are highly specific for target molecules.
- Immunoaffinity media – It uses antigens or antibodies (monoclonal or polyclonal) immobilized on matrix for isolation of specific targets.
- Immunoglobulin-binding protein media – These uses bacterial proteins binding to Fc region of antibodies. Examples is Protein A (from Staphylococcus aureus), Protein G (from Streptococci), Protein L.
- Lectin affinity media – It uses lectins binding carbohydrate residues for purification of glycoproteins and polysaccharides. Examples is Concanavalin A (Con A), Lentil Lectin, Wheat Germ Lectin.
- Avidin-biotin media – This exploits interaction between avidin (or streptavidin) and biotin for biotinylated substances.
- Nucleic acid affinity media – These uses immobilized DNA (DNA-cellulose) for purification of DNA-binding proteins like polymerases.
B. Pseudo-affinity media employs synthetic ligands.
It mimics natural substrates with broader specificity.
- Dye-ligand media – These uses textile dyes (Cibacron Blue F3G-A) as analogs for cofactors (NAD+/NADP+) or substrates. It is used for albumin and enzymes like kinases.
- Immobilized metal ion affinity chromatography (IMAC) media – It uses chelating ligands (IDA, NTA) with metal ions (Ni²⁺, Cu²⁺, Zn²⁺, Co²⁺) binding histidine or cysteine residues.
- Heparin media – This uses heparin (sulfated glycosaminoglycan) for binding DNA-binding proteins and coagulation factors.
C. Recombinant fusion protein media targets engineered proteins.
- Glutathione Sepharose – It binds proteins fused with Glutathione S-transferase (GST).
- Chelating Sepharose (IMAC) – These binds proteins with Poly-histidine tag (6xHis).
- Amylose resin – It binds proteins fused with Maltose Binding Protein (MBP).
- Streptavidin/Strep-Tactin – This binds proteins with Strep-tag.
- IgG Sepharose – It binds proteins fused with Protein A tag.
D. Small ligand/chemical media applies synthetic groups or inhibitors.
- Benzamidine media – These uses inhibitor for purification of serine proteases like trypsin.
- Boronate affinity media – It uses boronic acid binding cis-diol groups in nucleotides and glycated proteins (HbA1c).
- Amino acid resins – This uses amino acids (Arginine, Lysine) for binding proteins like plasminogen.
Types by Physical Support (Matrix)
A. Porous gel supports is used commonly.
These are beads for column packing.
- Agarose – It is widely used for low non-specific binding and hydrophilicity. Available in forms like Sepharose.
- Synthetic polymers – These includes polyacrylamide, polystyrene, polymethacrylate for mechanical strength.
- Dextran – Often combined with other materials.
- Silica/Glass – It is used in high-pressure applications but needs modification.
B. Alternative formats offers different handling.
- Magnetic beads – These is small particles coated with ligands for batch purification.
- Monoliths – It is continuous structures for high flow rates.
- Membranes – These is polymer or cellulose for rapid processing.
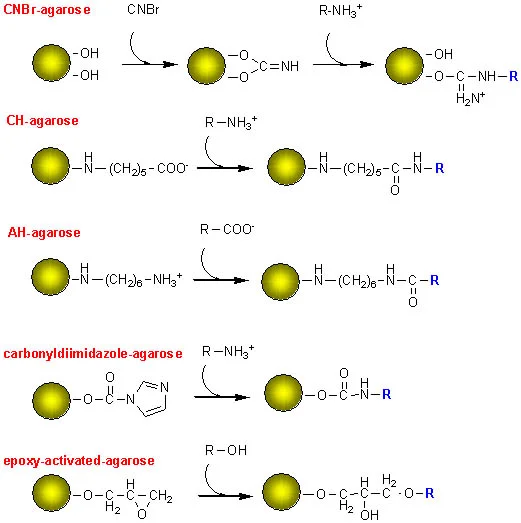
Pre-Activated Media
Pre-activated media allows coupling of custom ligands.
- NHS-activated – It is for ligands with primary amino groups.
- CNBr-activated – This is method for coupling proteins via amino groups.
- Epoxy-activated – It is for hydroxyl, amino, thiol groups.
- Thiol-activated – These is for reversible coupling of thiol proteins.
Application of Affinity Chromatography
- It is used for purification of recombinant proteins which are fused with affinity tags.
- It is used for purification of His-tagged proteins by using immobilized metal ions like Nickel (Ni²⁺) or Cobalt (Co²⁺).
- It is used for isolation of GST-tagged proteins with the help of glutathione immobilized on matrix.
- It is applied for purification of Maltose Binding Protein (MBP) fusion proteins using cross-linked amylose.
- It is used for separation of Strep-tagged proteins by using immobilized streptavidin or Strep-Tactin.
- It is also used for purification of proteins containing other tags such as FLAG tag, S-tag, CBP, and Halo-tag.
- It is used for purification of antibodies like IgG and its fragments by using Protein A, Protein G, and Protein L.
- It is applied for isolation of specific antibodies from serum or ascitic fluid by immobilizing antigen on the column.
- It is used for purification of specific antigens using immobilized monoclonal or polyclonal antibodies.
- It is used for purification of IgM and avian IgY using specific ligands or thiophilic media.
- It is used for purification of enzymes by immobilizing their specific substrates, inhibitors, or cofactors.
- It is applied for isolation of DNA-binding proteins such as polymerases and transcription factors using DNA-cellulose or heparin.
- It is used for separation of glycoproteins and polysaccharides using immobilized lectins like Concanavalin A.
- It is used for purification of blood coagulation factors using heparin affinity columns.
- It is applied for isolation of serine proteases such as trypsin and thrombin using benzamidine affinity matrix.
- It is used in clinical diagnosis for estimation of glycated hemoglobin (HbA1c) in blood.
- It is used for removal of interfering substances from patient samples before immunoassay tests.
- It is applied for detection of disease biomarkers in diagnostic and research studies.
- It is used for study of biomolecular interactions such as antigen–antibody interaction.
- It is applied for chiral separation of enantiomers using immobilized proteins as stationary phase.
- It is used in immunoprecipitation technique for isolation of specific proteins from cell lysate.
- It is applied in pull-down assays to study protein–protein interactions.
- It is used for removal of high-abundance proteins from serum samples during proteomic analysis.
- It is also used for separation of specific cell populations based on cell surface receptor binding.
Advantages of Affinity Chromatography
- It is a highly selective technique and gives very high resolution during separation of biomolecules.
- It can achieve a very high degree of purification in a single step.
- It is the only chromatographic technique which separates molecules based on their biological activity or specific chemical structure.
- It shows a concentrating effect as the target molecule gets concentrated during binding with the ligand.
- It allows processing of large volume of crude sample and the desired molecule is obtained in concentrated form.
- It saves time as extraction and purification can be carried out in a single step.
- It gives very high recovery of biologically active material.
- It is useful for separation of active molecules from denatured or inactive forms of the same substance.
- It is capable of isolating substances which are present in very low concentration in the sample.
- It is effective in removal of specific contaminants such as proteases from the sample.
- It allows reuse of affinity matrix for multiple purification cycles.
- It is generally not affected by sample volume as long as binding conditions and column capacity is maintained.
Disadvantages of Affinity Chromatography
- It is an expensive technique as affinity ligands are costly and sometimes not easily available.
- It requires large amount of solvents which increases the overall cost of the process.
- It is not economical for large scale purification due to high cost of bio-specific ligands.
- The procedure is labor intensive and requires skilled handling.
- Preparation of a specific affinity matrix takes more time when ready-made media is not available.
- Proteins may get denatured if proper pH is not maintained during binding or elution.
- Harsh elution conditions like low pH can damage the target molecule.
- Biological ligands such as antibodies are less stable compared to synthetic ligands.
- Some ligand–target interactions are very strong and elution becomes difficult.
- Non-specific binding cannot be completely eliminated during separation.
- Spacer arms may cause non-specific or irreversible adsorption of proteins.
- Leakage of ligand from the matrix can contaminate the purified product.
- In IMAC, metal ions may leach out at low pH and contaminate the sample.
- Metalloproteins are generally not suitable for IMAC as they bind the metal ions.
- Agarose matrices have low mechanical stability and cannot withstand high pressure.
- High ligand density may cause steric hindrance and reduce binding efficiency.
- Improper regeneration of column reduces binding capacity in repeated use.
- Reuse of column is limited and usually restricted to same type of sample.
Troubleshooting and Safety Considerations of Affinity Chromatography
The common problems encountered during affinity chromatography and their safety considerations are described below in proper list format–
Troubleshooting
- Target protein does not bind to column – The sample applied may not be in correct binding buffer. Improper pH or ionic strength reduces interaction between ligand and target. Inadequate equilibration of column also affects binding. Very fast flow rate reduces contact time and the column may be overloaded with excess sample.
- Poor or no elution of target protein– Very strong affinity between ligand and target prevents dissociation during elution. Elution buffer strength may be insufficient. Improper concentration of competitive ligand also results in incomplete elution. Sometimes broad peak is obtained due to slow dissociation of bound protein.
- Non-specific binding and contamination– High ligand density or unsuitable spacer arm may cause non-specific adsorption of proteins. Improper washing conditions allow contaminants to remain bound. Presence of lipids and lipoproteins in sample increases non-specific binding and reduces purity.
- Column clogging and pressure build-up– Turbid samples containing cell debris block the column bed. High lipid content in biological samples causes clogging. Air bubbles trapped inside column disturb flow and band separation.
Safety and Maintenance Considerations
- Protein stability during elution– Low pH elution buffers may denature proteins. Immediate neutralization of collected fractions is required. Protease activity may degrade target protein and protease inhibitors should be used when necessary.
- Use of chemicals– Sodium azide is toxic and interferes with coupling reactions. It must be handled carefully and removed before chromatography. Ethanol used for column storage is flammable and should be completely washed out before reuse.
- Ligand and metal ion leakage – Improper coupling or repeated use may result in ligand or metal ion leaching which contaminates purified product. This affects purity and biological activity.
- Column regeneration and reuse– Incomplete regeneration reduces binding capacity of column. Harsh cleaning buffers should be used carefully to avoid matrix damage. Reuse of column for different samples may cause cross contamination and should be avoided.
- General laboratory safety– Gloves lab coat and eye protection should be used while handling biological samples and chemicals. Proper waste disposal methods must be followed to prevent environmental and health hazards.
FAQ
Q1. What is affinity chromatography?
A. Affinity chromatography is a type of chromatographic technique which is used for separation and purification of biomolecules based on specific biological interaction. It depends on the specific binding between a target molecule and a ligand which is immobilized on stationary phase. This method is highly selective in nature and is widely used for purification of proteins enzymes and antibodies.
Q2. How does affinity chromatography work?
A. In affinity chromatography the stationary phase contains a ligand which has specific affinity towards the target molecule. When sample is passed through the column the desired molecule binds to ligand while other components are washed out. After binding the target molecule is eluted by changing pH ionic strength or by adding a competing ligand.
Q3. What are the applications of affinity chromatography?
A. Affinity chromatography is used for purification of enzymes antibodies hormones nucleic acid binding proteins and recombinant proteins. It is also used in biomedical research diagnostic laboratories and pharmaceutical industries. This technique is useful in studying protein–protein and protein–ligand interactions.
Q4. What are the advantages of affinity chromatography?
A. Some of the advantages are–
– High specificity and selectivity
– High purity of product is obtained in single step
– Suitable for purification of low concentration biomolecules
– Less time consuming compared to other methods
Q5. What are the different types of affinity chromatography?
A. The different types of affinity chromatography are–
– Immunoaffinity chromatography
– Dye ligand affinity chromatography
– Metal ion affinity chromatography (IMAC)
– Lectin affinity chromatography
– Avidin–biotin affinity chromatography
– Affinity chromatography using recombinant tags
Q6. How is affinity chromatography different from other chromatography methods?
A. Affinity chromatography is different from other chromatographic techniques because separation is based on specific biological interaction rather than physical properties like size charge or polarity. It provides very high selectivity compared to ion exchange gel filtration or adsorption chromatography.
Q7. What kind of interactions are exploited in affinity chromatography?
A. The interactions exploited include antigen–antibody interaction enzyme–substrate interaction receptor–ligand interaction metal ion–histidine interaction and lectin–carbohydrate interaction. These interactions are reversible and highly specific in nature.
Q8. What are the components of affinity chromatography?
A. The main components of affinity chromatography are–
– Stationary phase (matrix or support)
– Ligand
– Spacer arm
– Mobile phase (buffer)
– Elution system
Q9. What is the role of the ligand in affinity chromatography?
A. The ligand plays the major role in affinity chromatography. It is responsible for specific binding of target molecule. The ligand is immobilized on matrix and selectively binds the desired biomolecule from complex mixture.
Q10. How is elution achieved in affinity chromatography?
A. Elution in affinity chromatography is achieved by disrupting the interaction between ligand and target molecule. This is done by changing pH ionic strength temperature or by adding free ligand or chelating agents in case of metal affinity chromatography.
Q11. What is a spacer arm in affinity chromatography?
A. A spacer arm is a chemical group that connects ligand to matrix. It helps in reducing steric hindrance and improves accessibility of ligand for binding with target molecule. The spacer arm increases binding efficiency of the system.
Q12. What are common problems in affinity chromatography?
A. Some common problems are–
– Non specific binding of impurities
– Low binding capacity
– Ligand leakage from matrix
– Poor elution of target molecule
– Loss of biological activity of protein
Q13. How do you troubleshoot affinity chromatography?
A. Troubleshooting can be done by optimizing pH and buffer composition adjusting flow rate improving washing steps changing elution conditions and checking stability of ligand and target molecule. Proper regeneration of column also improves performance.
Q14. What are the key characteristics of affinity chromatography?
A. The key characteristics are high specificity high selectivity reversible interaction mild operating conditions and ability to purify biomolecules from complex mixtures in single step.
Q15. What are affinity tags in chromatography?
A. Affinity tags are short peptide or protein sequences attached to recombinant proteins to facilitate purification. These tags bind specifically to affinity media such as His-tag to metal ions GST-tag to glutathione and MBP-tag to amylose resin.
- Bickler, B. (2023, January 19). How to choose between linear gradients and step gradients for flash chromatography. Biotage. https://www.biotage.com/blog/how-to-choose-between-linear-gradients-and-step-gradients-for-flash-chromatography
- CallValuable6650. (2024). Can someone please explain this UGlobe question to me like I’m five? (I fear ChatGPT could not) [Online forum post]. Reddit. https://www.reddit.com/r/Mcat/comments/1iu97ar/can_someone_please_explain_this_uglobe_question/
- Chrom Tech. (2025, October 20). Understanding affinity chromatography. Chrom Tech, Inc. https://chromtech.com/blog/understanding-affinity-chromatography/
- GE Healthcare. (2007). Affinity chromatography: Principles and methods. General Electric Company.
- Hage, D. S. (1999). Affinity chromatography: A review of clinical applications. Clinical Chemistry, 45(5), 593–615. https://doi.org/10.1093/clinchem/45.5.593
- Hertz, C. M., Graves, D. J., Lauffenburger, D. A., & Serota, F. T. (1985). Use of cell affinity chromatography for separation of lymphocyte subpopulations. Biotechnology and Bioengineering, 27(5), 603–612. https://doi.org/10.1002/bit.260270509
- Lalli, E., Sarti, G. C., & Boi, C. (2018). Effect of the spacer arm on non-specific binding in membrane affinity chromatography. MRS Communications, 8(1), 65–70. https://doi.org/10.1557/mrc.2018.4
- Martínez, L. M. (2021, October 14). Affinity chromatography and column purification of proteins and nucleic acids. Sepmag. https://sepmag.eu/blog/affinity-chromatography-and-column-purification-of-proteins-and-nucleic-acids/
- Pina, A. S., Lowe, C. R., & Roque, A. C. A. (2014). Challenges and opportunities in the purification of recombinant tagged proteins. Biotechnology Advances, 32(2), 366–381. https://doi.org/10.1016/j.biotechadv.2013.12.001
- Rawal, A. (2025, April 3). Affinity chromatography: Principle, parts, steps, uses. Microbe Notes. https://microbenotes.com/affinity-chromatography/
- Rodriguez, E. L., Poddar, S., Iftekhar, S., Suh, K., Woolfork, A. G., Ovbude, S., Pekarek, A., Walters, M., Lott, S., & Hage, D. S. (2020). Affinity chromatography: A review of trends and developments over the past 50 years. Journal of Chromatography B, 1157, 122332. https://doi.org/10.1016/j.jchromb.2020.122332
- Sigma-Aldrich. (n.d.). Protein affinity chromatography. https://www.sigmaaldrich.com/US/en/technical-documents/technical-article/genomics/dna-and-rna-purification/affinity-chromatography
- Thermo Fisher Scientific. (n.d.). Overview of affinity purification. https://www.thermofisher.com/us/en/home/life-science/protein-biology/protein-biology-learning-center/protein-biology-resource-library/pierce-protein-methods/overview-affinity-purification.html
- Text Highlighting: Select any text in the post content to highlight it
- Text Annotation: Select text and add comments with annotations
- Comment Management: Edit or delete your own comments
- Highlight Management: Remove your own highlights
How to use: Simply select any text in the post content above, and you'll see annotation options. Login here or create an account to get started.