What is RNA?
- RNA, short for Ribonucleic acid, is a vital polymeric molecule that plays diverse roles in the biological processes of coding, decoding, regulation, and gene expression. Alongside DNA, lipids, proteins, and carbohydrates, RNA belongs to the nucleic acid family, which represents one of the fundamental macromolecules essential for all known life forms. While DNA forms a double helix structure, RNA exists as a single-stranded molecule that folds onto itself.
- In the realm of genetic information transfer, messenger RNA (mRNA) takes center stage. Cellular organisms rely on mRNA to convey genetic instructions using nitrogenous bases—guanine (G), uracil (U), adenine (A), and cytosine (C)—represented by the letters G, U, A, and C. This genetic information guides the synthesis of specific proteins. Intriguingly, many viruses utilize an RNA genome to encode their genetic information.
- Not limited to passive roles, certain RNA molecules actively participate in cellular processes by catalyzing biological reactions, controlling gene expression, and sensing and communicating cellular signals. Among these active processes, protein synthesis holds universal importance, wherein RNA molecules direct the synthesis of proteins on ribosomes. Transfer RNA (tRNA) molecules facilitate this process by delivering amino acids to the ribosome, while ribosomal RNA (rRNA) links these amino acids together to form coded proteins.
- Each RNA nucleotide comprises a ribose sugar with carbons numbered from 1′ to 5′. A base attaches to the 1′ position, typically adenine (A), cytosine (C), guanine (G), or uracil (U). Adenine and guanine belong to the purine category, whereas cytosine and uracil are pyrimidines. A phosphate group connects the 3′ position of one ribose and the 5′ position of the next, resulting in RNA’s negatively charged nature. Hydrogen bonds form between cytosine and guanine, adenine and uracil, and guanine and uracil. Additionally, other interactions, such as adenine bases binding together or the GNRA tetraloop formed by guanine–adenine base-pairs, are also possible.
- An important structural distinction between RNA and DNA lies in the presence of a hydroxyl group at the 2′ position of the ribose sugar. This group primarily leads RNA to adopt the A-form geometry, characterized by a deep and narrow major groove, as well as a shallow and wide minor groove. Furthermore, the 2′-hydroxyl group enables the chemical cleavage of the RNA backbone in flexible regions that are not involved in double helix formation.
- During RNA maturation, the four bases—adenine, cytosine, guanine, and uracil—can undergo various modifications. Notable examples include pseudouridine (Ψ) and ribothymidine (T), found in specific regions such as the TΨC loop of tRNA. Hypoxanthine, a deaminated adenine base, and its nucleoside form, inosine (I), also play essential roles in the wobble hypothesis of the genetic code. Moreover, there exist over 100 naturally occurring modified nucleosides, with tRNA showcasing the highest structural diversity of modifications, while rRNA commonly contains pseudouridine and nucleosides with 2′-O-methylribose.
- To fulfill their functional roles, single-stranded RNA molecules, similar to proteins, often require specific tertiary structures. Secondary structural elements, which encompass hydrogen bonds within the molecule, provide the scaffold for these structures. This results in recognizable “domains” of secondary structure, including hairpin loops, bulges, and internal loops. Designing RNA with a given secondary structure typically necessitates four bases, as fewer than four cannot create all structures, and more than four bases become unnecessary. Additionally, metal ions like Mg2+ are crucial for stabilizing many secondary and tertiary RNA structures due to the charged nature of RNA.
- The naturally occurring enantiomer of RNA is D-RNA, consisting of D-ribonucleotides with all chirality centers located in the D-ribose. L-RNA, synthesized using L-ribose or L-ribonucleotides, displays enhanced stability against degradation by RNase.
- Similar to proteins, the folded structure of an RNA molecule can possess a specific topology, which is defined based on the arrangement of intra-chain contacts within the folded RNA. This topological organization is commonly referred to as circuit topology.
- In summary, RNA encompasses various types that perform crucial functions in cellular processes. Understanding the types of RNA and their roles provides insights into the intricate mechanisms that govern genetic information transfer, gene expression, and protein synthesis.
Different Types of RNA and Their Functions
RNA, or Ribonucleic acid, is a crucial molecule synthesized by RNA polymerase from DNA. It serves various functions in the cell, including protein-coding and non-coding roles. Understanding the different types of RNA is essential for comprehending their specific functions. Let’s delve into the main types of RNA:
- Messenger RNA (mRNA): This type of RNA carries genetic information from DNA to the ribosomes, where proteins are synthesized. The sequences of mRNA determine the precise order of amino acids in the proteins produced.
- Ribosomal RNA (rRNA): RRNA plays a vital role in protein synthesis by incorporating into the ribosomes, the cellular structures responsible for assembling amino acids into proteins.
- Transfer RNA (tRNA): During translation, tRNA molecules are employed to transport specific amino acids to the growing polypeptide chains at the ribosomal site of protein synthesis. They ensure the accurate assembly of amino acids according to the mRNA code.
- Small nuclear RNA (snRNA): SnRNAs are involved in the processing and maturation of pre-mRNA, which is the initial transcript of protein-coding genes. They participate in the removal of introns and the splicing together of exons, ultimately forming the mature mRNA.
- MicroRNA (miRNA): MiRNAs are small RNA molecules, typically consisting of around 22 nucleotides. They play a crucial role in gene regulation by binding to specific messenger RNA (mRNA) molecules. This binding can result in the degradation of the targeted mRNA or inhibition of its translation into a protein.
- Small nucleolar RNA (snoRNA): SnoRNAs are involved in the modification and processing of other RNA molecules, particularly rRNA and tRNA. They guide specific chemical modifications, such as pseudouridylation and 2′-O-methylation, ensuring the functionality and stability of these RNA molecules.
- Long non-coding RNA (lncRNA): LncRNAs are RNA molecules that do not encode proteins but have diverse regulatory functions. They participate in gene expression regulation, chromatin remodeling, and other cellular processes, contributing to the overall complexity of cellular functions.
- Catalytic RNA (ribozymes): These RNA molecules possess enzymatic activity, functioning as catalysts within cells. They can perform various chemical reactions, such as RNA splicing or cleavage, without the need for protein assistance.
Messenger RNA (mRNA) – Structure and Functions
- Messenger ribonucleic acid (mRNA) is a single-stranded RNA molecule that plays a vital role in molecular biology by carrying the genetic information of a gene. This genetic sequence is then read by ribosomes during the process of protein synthesis.
- The creation of mRNA occurs through transcription, a process initiated by an enzyme called RNA polymerase. RNA polymerase converts the gene into a primary transcript known as pre-mRNA. Typically, pre-mRNA contains introns, which are non-coding regions that do not contribute to the final amino acid sequence. The removal of introns, called RNA splicing, results in the formation of mature mRNA, consisting only of exons that encode the protein sequence. This mature mRNA is then read by ribosomes.
- During translation, the ribosome utilizes transfer RNA (tRNA) to match the codons (sequences of three ribonucleotides) on the mRNA with specific amino acids. The tRNA molecules carry the corresponding amino acids, allowing the ribosome to assemble them in the correct order to form the protein. This process is an integral part of the central dogma of molecular biology, which describes the flow of genetic information within a biological system.
- The genetic information within mRNA is contained in the specific sequence of nucleotides, which form codons. Each codon codes for a specific amino acid, except for stop codons, which signal the termination of protein synthesis. The translation of codons into amino acids relies on two other types of RNA: transfer RNA (tRNA) and ribosomal RNA (rRNA). tRNA recognizes the codons on mRNA and delivers the corresponding amino acids, while rRNA constitutes the central component of the ribosome, the cellular machinery responsible for protein synthesis.
- The concept of mRNA was developed by Sydney Brenner and Francis Crick in 1960, during a conversation with François Jacob. In 1961, mRNA was independently identified and described by multiple teams, including one led by Brenner, Jacob, and Matthew Meselson, as well as another led by James Watson. During their data analysis in preparation for publication, Jacob and Jacques Monod coined the term “messenger RNA” to refer to this crucial molecule.
- In summary, messenger RNA (mRNA) acts as an intermediary in the transfer of genetic information from DNA to protein synthesis. Through transcription and RNA splicing, mRNA carries the coding information from the gene to the ribosome, where it is translated into a specific sequence of amino acids. The discovery and understanding of mRNA have revolutionized our comprehension of molecular biology and the fundamental processes governing life.
Structure of Messenger RNA (mRNA)
- mRNA was named by Jacob and Monad to signify its role as a messenger between DNA and proteins. It is a polymer made up of nucleotides, which are the building blocks of RNA. Each nucleotide consists of a phosphate group, a sugar molecule (ribose), and one of four nitrogenous bases: adenine (A), guanine (G), cytosine (C), and uracil (U). Notably, uracil replaces thymine (T) found in DNA.
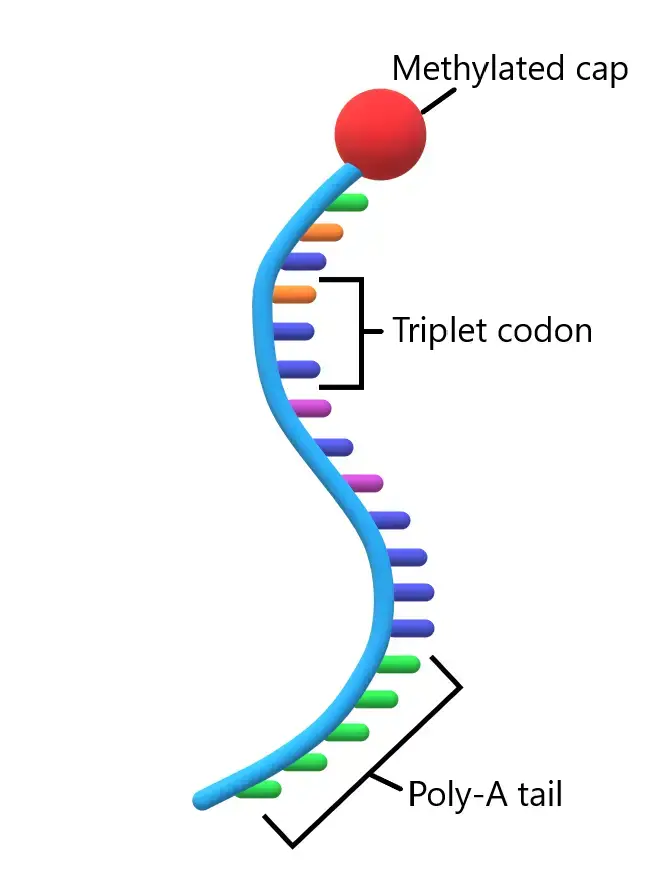
- The mature mRNA molecule can be divided into distinct regions, each playing a specific role in protein synthesis. These regions include the 5′ cap, 5′ untranslated region (UTR), coding region, 3′ UTR, and poly(A) tail.
- The coding region is composed of codons, which are triplets of nucleotides. Each codon corresponds to a specific amino acid, and ribosomes translate these codons into a polypeptide chain during protein synthesis. The coding region begins with the start codon ‘AUG’ and ends with one of the stop codons: UAG, UAA, or UGA. Additionally, certain segments of the coding sequence may possess regulatory functions.
- The untranslated regions (UTRs) are located before and after the coding region. The 5′ UTR precedes the start codon, while the 3′ UTR follows the stop codon. UTRs contribute to gene expression by influencing mRNA stability, translation efficiency, and mRNA localization within the cell.
- At the 5′ end of the mRNA molecule, there is a cap structure known as the 5′ cap. This cap consists of methyl guanosine triphosphate (m7G), which provides stability to the mRNA and assists in the recognition of the mRNA by ribosomes during translation.
- Conversely, at the 3′ end, there is a poly(A) tail composed of multiple adenine nucleotides. This polyadenylate tail serves various functions, such as protecting the mRNA from degradation, facilitating mRNA export from the nucleus, and enhancing translation efficiency.
- Depending on the organism and the specific mRNA, different characteristics can be observed. In eukaryotes, most mRNAs are monocistronic, meaning they code for a single protein. In contrast, prokaryotic mRNAs are often polycistronic, encoding multiple proteins. In such cases, the proteins typically have related functions and are regulated by a single regulatory region containing promoter and operator regions. Notably, the mitochondrial genome in humans is polycistronic.
Functions of Messenger RNA (mRNA)
- Messenger RNA (mRNA) plays a vital role in the process of protein synthesis within a cell. Its primary function is to serve as a template or messenger that carries the genetic information from the DNA in the nucleus to the ribosomes, where proteins are synthesized. This transfer of genetic information is known as transcription, and it is a crucial step in the central dogma of molecular biology.
- Once mRNA is transcribed from DNA, it undergoes a series of modifications, such as the addition of a 5′ cap and a poly-A tail, which helps protect it from degradation and facilitates its export from the nucleus to the cytoplasm. Once in the cytoplasm, mRNA serves as a blueprint for protein synthesis through a process called translation.
- During translation, the ribosomes bind to the mRNA and move along its length, reading the sequence of nucleotides in groups of three called codons. Each codon specifies a particular amino acid, which is the building block of proteins. The sequence of codons in the mRNA determines the order in which amino acids are assembled into a polypeptide chain, forming a specific protein.
- The genetic code is degenerate, meaning that multiple codons can code for the same amino acid. This redundancy provides flexibility and redundancy in protein synthesis. Additionally, the mRNA sequence contains start codons (usually AUG) and stop codons (such as UAA, UAG, or UGA), which mark the beginning and end of protein synthesis, respectively.
- In recent years, modified mRNA sequences have gained significant attention and have been utilized in the development of new therapeutic approaches and vaccines. Modified mRNA can be introduced into cells to produce specific proteins or trigger desired cellular responses. For example, in the context of vaccine development, mRNA vaccines work by introducing modified mRNA that encodes a viral antigen into cells. The cells then produce the antigen, which stimulates an immune response, leading to the production of antibodies and the development of immunity against the target pathogen.
- Prominent examples of mRNA vaccines include the Pfizer-BioNTech and Moderna vaccines, which have been developed and approved for restricted usage to protect against COVID-19. These vaccines have demonstrated high efficacy in preventing COVID-19 infection and have played a significant role in global vaccination efforts.
- In summary, the main functions of mRNA are to provide a template for protein synthesis, carry genetic information from the DNA to the ribosomes, and direct the cell to produce specific proteins. The use of modified mRNA sequences has shown great promise in various fields, including disease treatment and vaccine development, opening up new possibilities for targeted therapies and preventive measures.
Ribosomal RNA (rRNA) – Structure and Functions
- Ribosomal RNA (rRNA) is an essential component of ribosomes, which are the cellular structures responsible for protein synthesis in all cells. Unlike messenger RNA (mRNA) that carries genetic information from DNA to the ribosomes, rRNA is a type of non-coding RNA. It plays a crucial role in the translation process and acts as a ribozyme, an RNA molecule with enzymatic activity.
- The synthesis of rRNA occurs through transcription of ribosomal DNA (rDNA). Once transcribed, rRNA molecules bind to ribosomal proteins, forming the two subunits of the ribosome: the small subunit and the large subunit. These subunits work together to catalyze the assembly of amino acids into a polypeptide chain during protein synthesis.
- The physical and mechanical properties of rRNA within the ribosome are vital for the translation process. The ribosome provides a platform for the interaction of transfer RNA (tRNA) and mRNA. tRNA molecules bring amino acids to the ribosome, guided by the codons on the mRNA, while rRNA facilitates the binding of tRNA to the mRNA template. The rRNA in the ribosome acts as a catalyst, accelerating the formation of peptide bonds between adjacent amino acids and ensuring accurate protein synthesis.
- Interestingly, despite its critical role in protein synthesis, rRNA itself is not translated into proteins. Instead, it serves as a structural and functional component of the ribosome. In most cells, rRNA constitutes approximately 80% of the total cellular RNA, while the remaining 20% is composed of other RNA molecules, including mRNA and various non-coding RNAs.
- The ribosome is composed of approximately 60% rRNA and 40% ribosomal proteins by mass. This composition highlights the significant contribution of rRNA to the overall structure and function of ribosomes. The rRNA provides the scaffolding and catalytic framework for protein synthesis, while the ribosomal proteins contribute to the stability and regulation of the ribosome’s activity.
- In summary, ribosomal RNA (rRNA) is a non-coding RNA that plays a crucial role in protein synthesis. It forms the major component of ribosomes, which are responsible for translating genetic information into proteins. Through its interaction with ribosomal proteins, rRNA enables the accurate assembly of amino acids into polypeptide chains. Despite not being translated into proteins itself, rRNA is indispensable for the function and integrity of ribosomes, making it a fundamental component of all cells.
Ribosomal RNA (rRNA) Structure
- Ribosomal RNA (rRNA) possesses a specific structure that plays a crucial role in its function within ribosomes. Although the primary structure of rRNA can vary between organisms, it commonly forms stem-loop configurations through base-pairing. These stem-loops contribute to the formation of three-dimensional structures that are conserved across species. The rRNA stem-loops allow for specific interactions with ribosomal proteins, which are essential for the assembly of ribosomal subunits.
- The binding between rRNA and ribosomal proteins involves basic and aromatic residues present in the proteins. These residues form chemical interactions, such as stacking interactions, with the associated regions of rRNA. Additionally, ribosomal proteins can cross-link with the sugar-phosphate backbone of rRNA using basic residues. The specific sequences of ribosomal proteins that bind to rRNA have been identified. These interactions, along with the association of the small and large ribosomal subunits, lead to the formation of functional ribosomes capable of synthesizing proteins.
- Ribosomal RNA is organized into two major types of ribosomal subunits: the large subunit (LSU) and the small subunit (SSU). In prokaryotes, such as bacteria, the LSU is composed of a single small rRNA molecule and a single large rRNA molecule, combined with ribosomal proteins. The SSU contains a single small rRNA molecule. In eukaryotes, such as humans, the LSU consists of two small rRNA molecules and one large rRNA molecule, along with over 70 ribosomal proteins. The SSU contains a single small rRNA molecule.
- The rRNA sequences in both prokaryotes and eukaryotes are highly conserved, making them valuable for studying evolutionary relationships among organisms. The conservation of rRNA sequences over time is attributed to their vital role in ribosome function. In prokaryotes, the 16S rRNA is particularly used for phylogenetic analysis and delineation of species. The LSU rRNA is sometimes referred to as a ribozyme because ribosomal proteins do not bind to its catalytic site, known as the peptidyl transferase center. On the other hand, the SSU rRNA is involved in decoding mRNA in its decoding center, and ribosomal proteins cannot enter this region.
- The structure of rRNA is flexible and can undergo significant conformational changes that affect tRNA binding to the ribosome during translation. One example is the conformational switch observed in the 16S rRNA, where specific nucleotides alternate base pairing, altering the overall conformation of rRNA. This structural change can impact the ability of the ribosome to match a codon with its anticodon in tRNA selection and mRNA decoding.
- In summary, ribosomal RNA (rRNA) exhibits a specific structure characterized by stem-loop configurations, allowing for interactions with ribosomal proteins. These interactions contribute to the formation of ribosomal subunits and the overall function of the ribosome in protein synthesis. The conservation of rRNA sequences and its flexible structure play important roles in evolutionary studies and the regulation of translation processes.
Ribosomal RNA (rRNA) Functions
Ribosomal RNA (rRNA) serves several important functions in the process of protein synthesis. Here are some key functions of rRNA based on the provided information:
- Biogenesis of Ribosomes: rRNA is synthesized or transcribed in the cell nucleus, particularly in the nucleoli. The nucleoli play a significant role in the production of ribosomes by sequestering ribosomal proteins. The synthesis of rRNA and its subsequent assembly with ribosomal proteins are essential steps in the biogenesis of ribosomes.
- Formation of Ribosomal Subunits: Both prokaryotic and eukaryotic ribosomes are composed of two subunits, a larger subunit and a smaller subunit. During the translation of mRNA, these subunits come together to form a functional ribosome. In prokaryotes, the small subunit consists of a single RNA molecule approximately 1500 nucleotides long, known as the 16S rRNA, along with ribosomal proteins. The large subunit comprises two RNA molecules: one approximately 3000 nucleotides long (23S rRNA) and another short sequence of 120 nucleotides (5S rRNA), along with associated proteins.
- Subunit Sedimentation Rates: The small subunit of prokaryotic ribosomes, composed of the 16S rRNA and ribosomal proteins, has a sedimentation rate of 30S. The larger subunit, consisting of the 23S rRNA, 5S rRNA, and ribosomal proteins, has a sedimentation rate of 50S.
- Eukaryotic Ribosomal Subunits: Eukaryotic ribosomes also consist of two subunits, a large subunit (60S) and a small subunit (40S). The small subunit of eukaryotic ribosomes contains two short rRNA molecules, the 5S rRNA and the 5.8S rRNA, along with associated proteins. The large subunit is composed of two larger rRNA molecules: the 28S rRNA (over 5kb) and the 18S rRNA (2 kilobases). The 28S, 18S, and 5.8S rRNA molecules are generated through processing of a single primary transcript from a cluster of identical gene copies, while the 5S rRNA is produced from a separate cluster of identical genes. The eukaryotic ribosome has a sedimentation coefficient of 80S.
- Association with Organelles: In addition to the ribosomes present in the cytoplasm, eukaryotic cells also have rRNA in the mitochondria and chloroplasts. These organelles contain their own ribosomes, which play a crucial role in protein synthesis within these specific cellular compartments.
- Localization of Ribosomes: Ribosomes can be associated with the endoplasmic reticulum (ER) or exist as free-floating particles in the cytoplasm. Ribosomes associated with the ER are involved in synthesizing proteins that are destined for secretion or membrane insertion, while free ribosomes synthesize proteins that function within the cytoplasm.
Transfer RNA (tRNA) – Structure and Functions
Transfer RNA (tRNA) is a crucial molecule involved in protein synthesis, acting as an adaptor between messenger RNA (mRNA) and the amino acid sequence of proteins. Here are some key points about tRNA based on the provided information:
- Structure and Length: tRNA molecules are composed of RNA and typically range in length from 76 to 90 nucleotides in eukaryotes. The length of tRNA genes and mature tRNA varies across different organisms. Bacterial tRNAs are generally shorter, while eukaryotic tRNAs are shorter compared to Archaea.
- Role in Protein Synthesis: tRNA carries an amino acid to the ribosome, the cellular machinery responsible for protein synthesis. By complementing a three-nucleotide codon on mRNA with a three-nucleotide anticodon on tRNA, protein synthesis is achieved based on the mRNA code. Thus, tRNA plays a vital role in the process of translation.
- Anticodon and Genetic Code: The genetic code encoded in mRNA specifies which amino acids are incorporated into the protein product. tRNA molecules have an anticodon on one end that recognizes and binds to a specific codon on mRNA during protein biosynthesis. The anticodon forms complementary base pairs with the codon, ensuring accurate translation.
- Amino Acid Attachment: On the opposite end of the tRNA molecule, there is a covalent attachment to the amino acid corresponding to the anticodon sequence. Each tRNA molecule is specific to one type of amino acid, leading to the existence of multiple types of tRNA in an organism. Since multiple codons can code for the same amino acid, different tRNA molecules with different anticodons can carry the same amino acid.
- Aminoacyl tRNA Synthetases: The covalent attachment of amino acids to the 3′ end of tRNA is facilitated by enzymes called aminoacyl tRNA synthetases. These enzymes ensure that each tRNA molecule is correctly attached to its corresponding amino acid.
- Protein Synthesis Process: During protein synthesis, proteins called elongation factors aid in delivering tRNAs with attached amino acids to the ribosome. The tRNA interacts with the ribosome, facilitates the synthesis of the polypeptide chain, and participates in the translocation of the ribosome along the mRNA. If the anticodon of the tRNA matches the mRNA codon, the growing polypeptide chain is transferred to the newly delivered tRNA by another tRNA already bound to the ribosome.
- Chemical Modifications: Many nucleotides within tRNA can undergo chemical modifications, such as methylation or deamidation. These modifications can occur in various positions, including the anticodon region, and can influence the tRNA’s interaction with ribosomes and its base-pairing properties.
Transfer RNA (tRNA) Structure
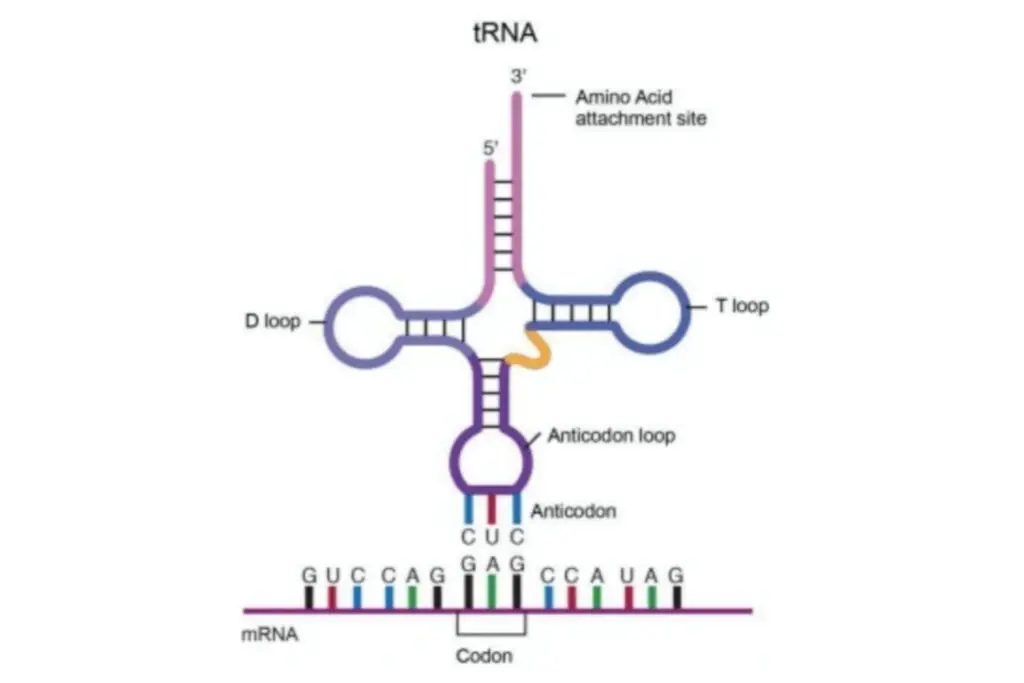
The structure of transfer RNA (tRNA) can be described in terms of its primary structure, secondary structure (often represented as a cloverleaf shape), and tertiary structure. These structural elements allow tRNA molecules to adopt a distinct L-shaped 3D structure that enables them to fit into specific sites on the ribosome during protein synthesis. Let’s explore the different components of tRNA structure:
- Acceptor Stem: The acceptor stem is a stem region composed of 7 to 9 base pairs. It forms through base pairing between the 5′-terminal nucleotide and the 3′-terminal nucleotide of the tRNA molecule. The 3′-terminal nucleotide also contains the CCA sequence, which is important for attaching the amino acid to the tRNA. The acceptor stem may include non-Watson-Crick base pairs.
- CCA Tail: The CCA tail is a sequence of cytosine-cytosine-adenine located at the 3′ end of the tRNA molecule. This sequence plays a crucial role in the recognition of tRNA by enzymes and is involved in translation. Aminoacyl tRNA synthetases covalently bond the amino acid to the 3′-hydroxyl group of the CCA tail. While prokaryotic tRNAs may have the CCA sequence transcribed in the tRNA gene, in most prokaryotic and eukaryotic tRNAs, the CCA sequence is added during tRNA processing.
- D Loop: The D loop is a stem-loop structure comprising 4 to 6 base pairs. It terminates in a loop region that often contains dihydrouridine. The D loop contributes to the overall stability and folding of the tRNA molecule.
- Anticodon Loop: The anticodon loop is another stem-loop structure consisting of 5 base pairs. Its loop region contains the anticodon sequence, which is crucial for recognizing and binding to the complementary codon on mRNA during protein synthesis. It is important to note that the primary structure of tRNA contains the anticodon in reverse order compared to the mRNA sequence because the reading of mRNA occurs in the 5′-to-3′ direction.
- ΨU Loop: The ΨU loop, named after the presence of the modified base ΨU (pseudo uridine), is found within a loop region of the tRNA. Pseudo uridine is a modified form of uridine. This loop, often appearing as 5′ -TΨUCG-3′, contributes to the overall structure and function of tRNA.
- Variable Loop: Situated between the anticodon loop and the ΨU loop, the variable loop is aptly named as it varies in size across different tRNA molecules. It can range from 3 to 21 bases and contributes to the diversity and flexibility of tRNA structures.
Transfer RNA (tRNA) Functions
Transfer RNA (tRNA) plays a crucial role in protein synthesis by serving as an adapter molecule that connects the genetic information encoded in mRNA with the corresponding amino acid sequence in proteins. Here are the key functions of tRNA:
- Amino Acid Carriers: The primary function of tRNA is to carry amino acids to the ribosome during protein synthesis. Each tRNA molecule is specifically bound to a particular amino acid by aminoacyl-tRNA synthetases, enzymes that catalyze the attachment of amino acids to the tRNA molecules. This specific coupling ensures that the correct amino acid is incorporated into the growing polypeptide chain.
- Recognition of mRNA Codons: tRNA possesses an anticodon sequence that is complementary to the codon sequence on mRNA. The anticodon is located within the anticodon loop of the tRNA molecule. During translation, the anticodon base pairs with the corresponding codon on mRNA, ensuring the accurate selection of the amino acid to be added to the growing polypeptide chain.
- Translocation of Amino Acids: tRNA participates in the translocation process during protein synthesis. Once a tRNA molecule delivers its amino acid to the ribosome, it moves from the A (aminoacyl) site to the P (peptidyl) site and then to the E (exit) site of the ribosome. This movement is facilitated by various elongation factors and ensures the correct order and sequence of amino acids in the growing polypeptide chain.
- Ribosome Binding: tRNA molecules interact with ribosomes, the cellular machinery responsible for protein synthesis. The L-shaped structure of tRNA allows it to fit into the ribosome’s P and A sites, ensuring the proper alignment and positioning of the mRNA codon and the corresponding tRNA anticodon for accurate translation.
- Fine-Tuning of Translation: Some tRNA molecules contain modified nucleotides, such as pseudouridine or methylated bases. These modifications can influence the stability of tRNA structure, its interaction with the ribosome, and the decoding accuracy during translation. Additionally, modified nucleotides within the anticodon region of certain tRNAs can enhance base-pairing flexibility, enabling recognition of multiple codons for a specific amino acid.
Small Nuclear RNA (snRNA) – Structure and Functions
Small nuclear RNA (snRNA) is a class of RNA molecules with various functions in the nucleus of eukaryotic cells. Here are the key aspects of snRNA structure and its functions:
Small Nuclear RNA (snRNA) Structure
- Length and Transcription: snRNA molecules are approximately 150 nucleotides long and are transcribed by either RNA polymerase II or RNA polymerase III.
- Association with Proteins: snRNAs are always associated with specific proteins, forming complexes called small nuclear ribonucleoproteins (snRNPs). These complexes consist of snRNA and snRNP-specific proteins, such as the Sm proteins. Common snRNAs in human cells include U1, U2, U4, U5, and U6 spliceosomal RNAs.
Small Nuclear RNA (snRNA) Functions
- Pre-mRNA Processing: The primary function of snRNAs is to participate in the processing of pre-messenger RNA (hnRNA) in the nucleus. Specifically, snRNAs are involved in the splicing of introns and the excision of introns to produce mature mRNA. They are part of the spliceosome, a large complex responsible for the precise removal of introns and the joining of exons.
- Transcriptional Regulation: Certain snRNAs, such as 7SK RNA and B2 RNA, have been shown to regulate transcription factors or RNA polymerase II activity. These snRNAs can influence gene expression by modulating the function of these regulatory elements.
- Telomere Maintenance: Some snRNAs play a role in maintaining telomeres, the protective caps at the ends of chromosomes. Telomeres are essential for chromosome stability and replication. snRNAs are involved in the complex machinery that ensures the proper length and function of telomeres.
- Association with Cajal Bodies: snRNAs are found within splicing speckles and Cajal bodies in the nucleus. Cajal bodies are specialized compartments involved in the biogenesis of small nuclear ribonucleoproteins (snRNPs) and small nucleolar RNAs (snoRNAs). They serve as sites for snRNP assembly and modification.
- Differentiation from snoRNAs: snRNAs should not be confused with small nucleolar RNAs (snoRNAs), which guide chemical modifications of ribosomal RNAs (rRNAs) and other RNA genes. Although both snRNAs and snoRNAs are small RNA molecules involved in RNA biogenesis, they have distinct functions and are classified under the broader category of small RNAs.
In summary, snRNA plays a crucial role in pre-mRNA processing, participating in the splicing of introns and the formation of mature mRNA. They also have additional functions in transcriptional regulation, telomere maintenance, and association with specialized nuclear structures. The formation of snRNPs with specific proteins ensures their proper functioning and stability within the nucleus.
Long Non-coding RNA (lncRNA) – Structure and Functions
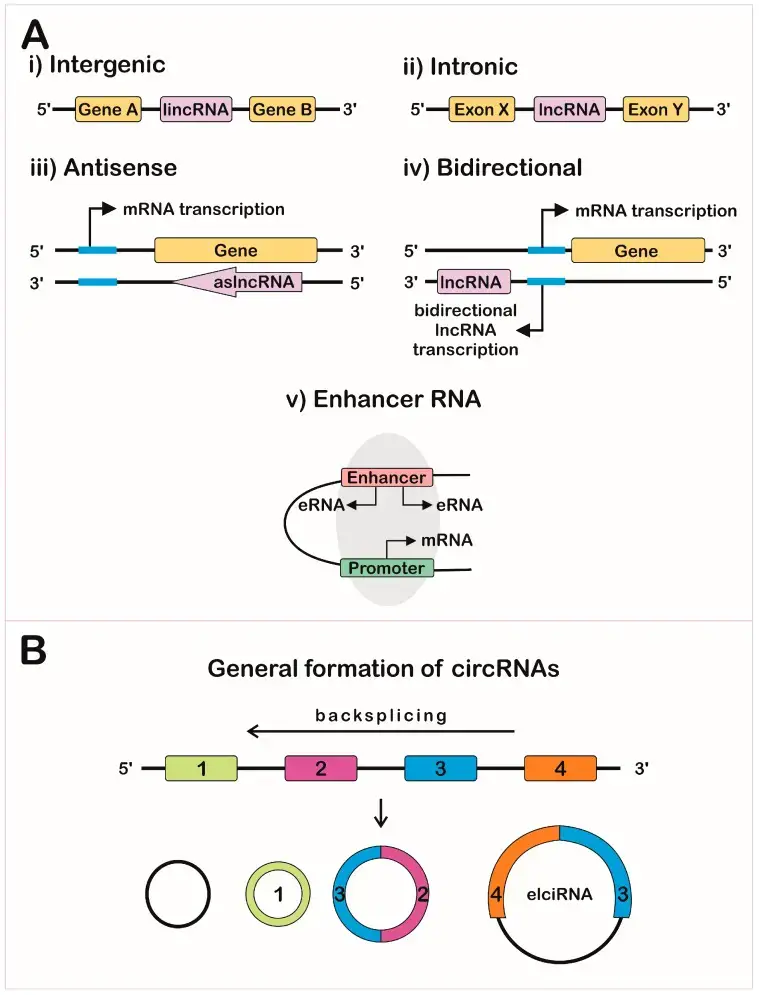
- Long non-coding RNAs (lncRNAs) are a diverse class of RNA molecules that play important regulatory roles in various cellular processes. Unlike messenger RNAs (mRNAs) that code for proteins, lncRNAs do not encode proteins themselves. They are generally defined as RNA transcripts longer than 200 nucleotides. This length criterion distinguishes them from small non-coding RNAs, such as microRNAs and small interfering RNAs, which have distinct functions in gene regulation.
- LncRNAs exhibit different genomic positions and can be classified into several subtypes. These include intergenic lncRNAs, which are located between protein-coding genes; intronic lncRNAs, which are derived from introns of protein-coding genes; and sense and antisense lncRNAs, which overlap with protein-coding genes in the same or opposite direction.
- Research conducted in 2007 indicated that only a small fraction of transcription across the human genome is associated with protein-coding genes, suggesting the existence of a vast amount of long non-coding RNA sequences. Large-scale sequencing projects, such as the FANTOM (Functional Annotation of Mammalian Genomes) project, have provided valuable insights into the complexity of lncRNA transcription. For instance, the FANTOM3 project identified tens of thousands of non-coding transcripts that possess characteristics similar to mRNAs, such as 5′ capping, splicing, and polyadenylation. However, these transcripts lack an open reading frame (ORF) for protein synthesis.
- Identifying lncRNAs is a challenging task due to the difficulty in distinguishing them from protein-coding transcripts. Nevertheless, multiple studies have shown that the testis and neural tissues express a significant amount of lncRNAs compared to other tissue types. The FANTOM5 project, utilizing advanced techniques, has identified thousands of lncRNAs in various human sources, totaling to 27,919 long non-coding RNAs.
- In terms of abundance, lncRNAs are typically expressed at lower levels compared to mRNAs. This lower abundance is attributed to the higher cell-to-cell variation in expression levels of lncRNA genes compared to protein-coding genes. Moreover, lncRNAs exhibit higher tissue specificity and developmental stage specificity than mRNAs. They play critical roles in various biological processes, including brain development and function.
- While the majority of studies on lncRNAs have focused on mammals, recent efforts have also explored their prevalence in plants. A comprehensive study involving multiple plant species identified a significant number of non-coding transcripts, leading to the establishment of the Green Non-Coding Database (GreeNC), a repository of plant lncRNAs.
- The genomic organization of lncRNAs is complex, with many lncRNAs overlapping with protein-coding genes. This overlap often results in a hierarchy of overlapping isoforms, adding to the complexity of lncRNA research. The GENCODE consortium has made significant contributions to the annotation and analysis of human lncRNAs, providing insights into their genomic organization, modifications, cellular locations, and tissue expression profiles. It has been observed that human lncRNAs exhibit a bias toward two-exon transcripts.
- In conclusion, long non-coding RNAs represent a diverse and important class of regulatory molecules that contribute to gene regulation and various cellular processes. They exhibit unique characteristics, including tissue specificity and developmental stage specificity. While the study of lncRNAs is still evolving, their significance in both mammals and plants is increasingly recognized, and ongoing research continues to unravel their roles in gene regulation and cellular functions.
Long Non-coding RNA (lncRNA) Structure
The structure of long non-coding RNAs (lncRNAs) can vary widely, and their diverse structures contribute to their functional versatility. Unlike messenger RNAs (mRNAs) that have a well-defined structure consisting of a coding region and untranslated regions (UTRs), lncRNAs lack a consistent structure due to their non-coding nature. However, certain structural features are commonly observed in lncRNAs.
- Linear Structure: LncRNAs, like other RNA molecules, have a linear structure consisting of a sequence of nucleotides. These nucleotides can be adenine (A), cytosine (C), guanine (G), or uracil (U) in the case of RNA, as opposed to thymine (T) in DNA. The sequence of nucleotides in an lncRNA molecule determines its functional properties and interactions with other molecules.
- Secondary Structure: LncRNAs can adopt various secondary structures, including stem-loops, hairpins, bulges, internal loops, and pseudoknots. These structural motifs are formed by base pairing between complementary nucleotides within the lncRNA sequence. Secondary structures contribute to the stability and functional properties of lncRNAs, such as their ability to interact with proteins or other RNA molecules.
- Higher-Order Structure: In addition to secondary structures, lncRNAs can form complex three-dimensional structures. The folding of lncRNAs into higher-order structures is often mediated by interactions between different regions of the molecule, involving base pairing, stacking interactions, and tertiary interactions. The specific folding pattern of an lncRNA contributes to its overall structure and function.
- Domains and Motifs: LncRNAs can contain specific domains or motifs within their sequences, which are responsible for their functional interactions. These domains may include protein-binding domains, RNA-binding domains, or specific structural motifs associated with regulatory functions. Domains within lncRNAs allow them to interact with other molecules, including proteins, DNA, or other RNA molecules, to carry out their regulatory roles.
- Chromatin Structure: Some lncRNAs play a role in modulating chromatin structure and gene expression. These lncRNAs can interact with chromatin and form complexes with chromatin-modifying proteins, contributing to the regulation of gene expression. The chromatin-associated lncRNAs may have specific structural features that facilitate their interactions with chromatin and associated proteins.
Long Non-coding RNA (lncRNA) Functions
- Long non-coding RNAs (lncRNAs) have been found to have various functions in the regulation of gene expression. While only a small proportion of lncRNAs have been extensively studied and functionally annotated, accumulating evidence suggests that the majority of lncRNAs are likely to play important roles in cellular processes.
- One major function of lncRNAs is in the regulation of gene transcription. They can act as co-regulators, modifying the activity of transcription factors or regulating the association and activity of co-regulators. For example, lncRNA Evf-2 acts as a co-activator for the homeobox transcription factor Dlx2, which is involved in forebrain development and neurogenesis. Another example is the role of lncRNAs in recruiting transcriptional programs to regulate adjacent protein-coding gene expression. Divergent lncRNAs that are transcribed in the opposite direction to nearby protein-coding genes have been shown to regulate the transcription of nearby essential developmental regulatory genes.
- Furthermore, lncRNAs can target the basal transcription machinery, which includes general transcription factors required for the RNA polymerase II (RNAP II) transcription of all genes. They can affect promoter usage and control gene expression by forming stable RNA-DNA triplexes or modulating the activity of transcription factors involved in transcription initiation or elongation. Additionally, lncRNAs transcribed by RNA polymerase III (RNAP III) can regulate the expression of RNAP II-dependent genes, forming a functional regulatory network between these two RNA polymerases.
- In addition to their roles in transcriptional regulation, lncRNAs also participate in post-transcriptional gene regulation. They can control various aspects of mRNA processing, such as splicing, transport, translation, and degradation. Through complementary base pairing with target mRNAs, lncRNAs can influence their fate and function. For example, lncRNAs can regulate alternative splicing of specific mRNAs or control their translation in response to synaptic activity in neurons. They can also form double-stranded RNA duplexes, leading to the generation of endogenous small interfering RNAs (siRNAs) that can regulate gene expression.
- Another important function of lncRNAs is in epigenetic regulation. They can interact with chromatin and recruit chromatin-modifying complexes to specific genomic loci, thereby influencing the epigenetic state and gene expression patterns. Examples include lncRNA HOTAIR, which recruits Polycomb chromatin remodeling complexes to repress transcription across large chromatin domains, and lncRNA CDKN2BAS, which can induce changes to heterochromatin formation and affect gene expression.
- In summary, lncRNAs have diverse functions in the regulation of gene expression. They can act at multiple levels, including transcriptional regulation, post-transcriptional regulation, and epigenetic regulation. Although the functional annotation of lncRNAs is still an ongoing area of research, their importance in cellular processes is increasingly being recognized.
Small Nucleolar RNA (snoRNA) – Structure and Functions
- Small nucleolar RNAs (snoRNAs) are a class of small RNA molecules ranging from approximately 60 to 300 nucleotides in length. They are primarily located in the nucleolus, a subnuclear organelle responsible for the synthesis and assembly of ribosomes. SnoRNAs have been found to serve various essential functions within the cell, contributing to processes such as ribosome synthesis, chemical modifications of RNA molecules, splicing of pre-mRNA, and even telomere synthesis.
- One of the key roles of snoRNAs is their involvement in ribosome synthesis. Ribosomes are the cellular machinery responsible for protein synthesis. SnoRNAs contribute to this process by cleaving and processing the large precursor RNA molecules that give rise to the mature 28S, 18S, and 5.8S rRNAs, which are integral components of ribosomes. Through precise cleavage and modification of these precursor molecules, snoRNAs ensure the production of functional rRNAs necessary for ribosome assembly.
- Furthermore, snoRNAs are also responsible for chemically modifying specific nucleotides in a variety of RNA molecules, including rRNA, tRNA, and snRNA. These modifications involve the addition of various chemical groups, such as methyl groups, to the ribose backbone of the RNA molecule. These chemical modifications play crucial roles in fine-tuning the structure, stability, and function of RNA molecules. By adding methyl groups, snoRNAs contribute to the regulation and optimization of RNA activity, ultimately affecting protein synthesis and other cellular processes.
- Another important function of snoRNAs is their involvement in pre-mRNA splicing. Pre-mRNA molecules undergo extensive processing steps before becoming mature mRNA molecules that can be translated into proteins. SnoRNAs assist in this process by aiding in the splicing of pre-mRNA into different forms of mature mRNA. They contribute to the precise removal of introns, non-coding regions within pre-mRNA, and facilitate the precise stitching together of exons, the coding regions, to generate functional mRNA molecules.
- Interestingly, certain types of snoRNAs also serve as templates for the synthesis of telomeres, the protective structures located at the ends of chromosomes. Telomeres play a crucial role in maintaining genomic stability and preventing the loss of genetic information during DNA replication. SnoRNAs act as a template during the synthesis of telomeres, ensuring the proper replication and preservation of chromosome ends, thus contributing to the overall integrity of the genome.
- In vertebrates, snoRNAs are primarily derived from introns, which are non-coding regions within genes that are removed during the transcription process. The introns are usually discarded as waste material, but in the case of snoRNAs, they are repurposed to generate functional RNA molecules with important cellular roles. This recycling of intronic regions highlights the efficiency and versatility of the cellular machinery in utilizing available resources.
- In summary, small nucleolar RNAs (snoRNAs) are a diverse group of small RNA molecules found in the nucleolus. They perform various crucial functions within the cell, including ribosome synthesis, chemical modification of RNA molecules, pre-mRNA splicing, and even telomere synthesis. Through their involvement in these processes, snoRNAs contribute to the regulation of gene expression, protein synthesis, and the overall stability and functionality of the genome.
MicroRNAs (miRNAs) – Structure and Functions
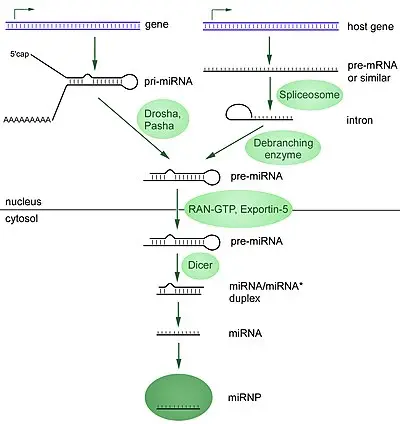
- MicroRNAs (miRNAs) are a class of small, single-stranded, non-coding RNA molecules consisting of approximately 22 nucleotides. They share a similar size with another class of small RNA molecules called small interfering RNAs (siRNAs). MiRNAs are present in plants, animals, and certain viruses, and they play a crucial role in RNA silencing and the regulation of gene expression at the post-transcriptional level.
- In humans, it is estimated that around 1000 miRNAs are generated. These miRNAs can be encoded in the genome either as stand-alone genes or within introns of other genes. Regardless of their location, miRNAs serve as important regulators of mRNA within cells. Their expression is often specific to certain cell types and can vary during the differentiation of these cell types.
- The primary function of miRNAs is to regulate gene expression by interacting with mRNA molecules. They achieve this in two distinct ways, depending on the degree of sequence complementarity between the miRNA and the target mRNA. When the sequences are evenly matched, particularly in plants, miRNAs can lead to the destruction of the target mRNA. On the other hand, when the sequences are only partially matched, miRNAs repress the translation of the target mRNA, preventing it from being efficiently translated into protein.
- These characteristic roles of miRNAs can be attributed to two key features. Firstly, their small size enables rapid transcription from their respective genes. This allows for the production of numerous miRNAs, which can quickly influence gene expression within cells. Secondly, miRNAs do not require a protein component for their regulatory function. Unlike some other gene regulators, miRNAs can directly interact with mRNA and modulate its expression without the need for additional translation factors.
- Genomic studies have shown that one or more miRNAs can bind to specific mRNA molecules transcribed from DNA. It has been observed that a single miRNA can bind to approximately 200 different mRNA targets. This is facilitated by the presence of multiple miRNA binding sites on the mRNA molecules. By binding to multiple targets, miRNAs enable coordinated regulation of mRNA translation, leading to the fine-tuning of gene expression in cells.
- In summary, microRNAs (miRNAs) are small, non-coding RNA molecules that play a crucial role in post-transcriptional gene expression regulation. They are approximately 22 nucleotides in length and are found in plants, animals, and some viruses. MiRNAs regulate gene expression by interacting with mRNA molecules, either by causing mRNA degradation or by repressing translation. Their small size and protein-independent regulatory mechanism make them efficient and versatile regulators of gene expression. By binding to multiple mRNA targets, miRNAs enable coordinated mRNA translation, contributing to the precise control of gene expression within cells.
Catalytic RNA (ribozymes) – Structure and Functions
- Ribozymes, also known as ribonucleic acid enzymes, are RNA molecules that possess catalytic activity and can facilitate specific biochemical reactions. This discovery in 1982 demonstrated that RNA can function both as genetic material, like DNA, and as a biological catalyst, akin to protein enzymes. The existence of ribozymes supported the RNA world hypothesis, which suggests that RNA may have played a crucial role in the evolution of self-replicating systems in the early stages of life.
- Ribozymes exhibit various catalytic activities, including cleavage (or ligation) of RNA and DNA, as well as peptide bond formation. For example, the smallest known ribozyme, GUGGC-3′, is capable of aminoacylating a GCCU-3′ sequence in the presence of PheAMP. Within the ribosome, ribozymes function as part of the large subunit ribosomal RNA, facilitating the linking of amino acids during protein synthesis. Additionally, ribozymes are involved in numerous RNA processing reactions, such as RNA splicing, viral replication, and transfer RNA biosynthesis. Examples of ribozymes include the hammerhead ribozyme, the VS ribozyme, leadzyme, and the hairpin ribozyme.
- Researchers investigating the origins of life through the RNA world hypothesis have been striving to discover a ribozyme capable of self-replication. Such a ribozyme would need to possess the ability to catalytically synthesize RNA polymers under prebiotically plausible conditions. This process should occur with high accuracy to prevent information degradation, while still allowing occasional errors during copying to enable Darwinian evolution.
- Ribozymes have also been explored as potential therapeutic agents, particularly in targeting specific RNA sequences for cleavage. They hold promise for applications in gene therapy, biosensors, functional genomics, and gene discovery. Modifying the 2′ position on the ribose of catalytic RNA molecules has been a strategy to enhance their stability, as ribozymes have relatively short half-lives in the body. One area of research in ribozyme gene therapy focuses on inhibiting RNA-based viruses. Synthetic ribozymes, such as gene shears, have been developed and entered clinical testing for HIV infection. Ribozymes have also been designed to target other viruses such as hepatitis C, SARS coronavirus, adenovirus, and influenza A and B viruses. These ribozymes can cleave conserved regions of the viral genome and have shown efficacy in reducing viral levels in cell cultures. However, further development and clinical translation of ribozyme-based therapies are still ongoing.
FAQ
What is messenger RNA (mRNA)?
Messenger RNA (mRNA) is a type of RNA molecule that carries genetic information from the DNA in the nucleus to the ribosomes in the cytoplasm, where it serves as a template for protein synthesis.
What is transfer RNA (tRNA)?
Transfer RNA (tRNA) is a type of RNA molecule that plays a crucial role in protein synthesis. It binds to specific amino acids and delivers them to the ribosome, where they are incorporated into the growing polypeptide chain.
What is ribosomal RNA (rRNA)?
Ribosomal RNA (rRNA) is a type of RNA molecule that forms the structural and catalytic core of the ribosome. It helps in the assembly of proteins by decoding the information carried by mRNA and catalyzing the formation of peptide bonds.
What is small nuclear RNA (snRNA)?
Small nuclear RNA (snRNA) is a type of RNA molecule found in the nucleus of eukaryotic cells. It plays a crucial role in pre-mRNA splicing, which is the process of removing introns and joining exons to produce mature mRNA molecules.
What is microRNA (miRNA)?
MicroRNA (miRNA) is a small non-coding RNA molecule involved in post-transcriptional gene regulation. It binds to specific mRNA molecules and inhibits their translation or promotes their degradation, thereby regulating gene expression.
What is small interfering RNA (siRNA)?
Small interfering RNA (siRNA) is a type of RNA molecule that regulates gene expression by silencing specific genes. It is typically introduced into cells or organisms to induce RNA interference (RNAi) and suppress the expression of targeted genes.
What is long non-coding RNA (lncRNA)?
Long non-coding RNA (lncRNA) is a type of RNA molecule that does not code for proteins. It is involved in various cellular processes, such as chromatin modification, gene regulation, and protein localization.
What is circular RNA (circRNA)?
Circular RNA (circRNA) is a type of RNA molecule that forms a closed loop structure. It is generated through a process called back-splicing and has been found to have regulatory roles in gene expression and cellular processes.
What is heterogeneous nuclear RNA (hnRNA)?
Heterogeneous nuclear RNA (hnRNA) refers to the primary transcripts of protein-coding genes before they undergo processing, including capping, splicing, and polyadenylation, to become mature mRNA molecules.
What is viral RNA?
Viral RNA refers to the RNA genome of viruses. Different types of viruses can have different RNA genomes, including single-stranded RNA (ssRNA), double-stranded RNA (dsRNA), and retroviral RNA, among others.
References
- Cech, T. R. (2014). The RNA Worlds in Context. Cold Spring Harbor Perspectives in Biology, 6(11), a016742. doi: 10.1101/cshperspect.a016742
- Gilbert, W. V., & Bell, T. A. (2019). Messenger RNA modifications: Form, distribution, and function. Science, 365(6451), eaau1765. doi: 10.1126/science.aau1765
- Noller, H. F. (2005). RNA Structure: Reading the Ribosome. Science, 309(5740), 1508-1514. doi: 10.1126/science.1115622
- Hüttenhofer, A., Schattner, P., & Polacek, N. (2005). Non-coding RNAs: Hope or Hype? Trends in Genetics, 21(5), 289-297. doi: 10.1016/j.tig.2005.03.007
- Bartel, D. P. (2009). MicroRNAs: Target Recognition and Regulatory Functions. Cell, 136(2), 215-233. doi: 10.1016/j.cell.2009.01.002
- Breaker, R. R. (2012). Riboswitches and the RNA World. Cold Spring Harbor Perspectives in Biology, 4(2), a003566. doi: 10.1101/cshperspect.a003566
- Ferré-D’Amaré, A. R., & Scott, W. G. (2010). Small Self-Cleaving Ribozymes. Cold Spring Harbor Perspectives in Biology, 2(5), a003574. doi: 10.1101/cshperspect.a003574
- Saleh, M.-C., van Rij, R. P., Hekele, A., Gillis, A., Foley, E., O’Farrell, P. H., … Andino, R. (2006). The Endocytic Pathway Mediates Cell Entry of dsRNA to Induce RNAi Silencing. Nature Cell Biology, 8(8), 793-802. doi: 10.1038/ncb1439
- Wang, Q., Lee, I., Ren, J., Ajuyah, P., Lee, Y.-L., & Bao, X. (2020). Ribozymes: Functions and Mechanisms in RNA Regulation. Genes, 11(6), 631. doi: 10.3390/genes11060631
- Roth, A., & Breaker, R. R. (2009). The Structural and Functional Diversity of Small Molecule RNA Regulators. Cell, 136(3), 515-529. doi: 10.1016/j.cell.2009.01.043
- Holbrook, S. R. (2018). RNA Structure: The Long and the Short of It. Current Opinion in Structural Biology, 49, 79-85. doi: 10.1016/j.sbi.2018.01.013
- Bartel, D. P. (2018). Metazoan MicroRNAs. Cell, 173(1), 20-51. doi: 10.1016/j.cell.2018.03.006
- Krol, J., & Krzyzosiak, W. J. (2006). Structure and functions of 3′ untranslated regions of mammalian messenger RNAs. Acta Biochimica Polonica, 53(4), 693-707.