What is Two-Point Cross?
A two-point cross is a genetic mapping technique used to determine the recombination frequency between two specific genes located on the same chromosome. In this method, a test cross is performed between a heterozygous individual (carrying two different alleles for the genes of interest) and a homozygous recessive individual (which has two recessive alleles for those genes).
The primary goal of a two-point cross is to calculate the frequency of recombination events that occur between the two genes during meiosis. This frequency can then be used to estimate the genetic distance between the two genes on the chromosome. The closer the genes are to each other, the lower the recombination frequency, as they are more likely to be inherited together. Conversely, genes that are farther apart are more likely to be separated by recombination events.
In summary, a two-point cross helps geneticists understand the linkage and relative positions of genes on a chromosome by analyzing the offspring produced from the test cross.
What is the purpose of a two-point cross in genetics?
The purpose of a two-point cross in genetics is to determine the linkage between two genes and to calculate the recombination frequency between them. This method allows geneticists to:
- Assess Gene Linkage: By analyzing the offspring produced from a cross between a homozygous recessive individual and a heterozygous individual for two genes, researchers can determine whether the genes are linked (located close together on the same chromosome) or assort independently.
- Calculate Recombination Frequency: The two-point cross provides data on the frequency of recombination events that occur between the two genes. This frequency is used to estimate the genetic distance between the genes, which is expressed in map units (centimorgans).
- Create Genetic Maps: The information obtained from multiple two-point crosses can be used to construct genetic maps, which visually represent the relative positions of genes on a chromosome.
- Study Inheritance Patterns: Two-point crosses help in understanding how traits are inherited together, which can be important for studying genetic diseases and traits in various organisms.
Methodology used in conducting a Two-point cross
A two-point cross is a genetic cross used to determine the linkage and relative distance between two genes located on the same chromosome. Here’s a step-by-step guide to the methodology:
- Select Parent Organisms: Choose two parent organisms that are homozygous for different alleles of the two genes you want to study. For example, if you’re studying genes A and B, one parent might be homozygous for alleles A1A1 and B1B1, and the other parent might be homozygous for alleles A2A2 and B2B2.
- Perform the Cross: Cross these two parents to produce an F1 generation that is heterozygous for both genes (A1A2, B1B2).
- Create a Test Cross: Cross the F1 individuals (A1A2, B1B2) with a tester organism that is homozygous recessive for both genes (A1A1, B1B1). This cross is known as a test cross and helps reveal the genotype of the F1 individuals.
- Analyze the Offspring: Examine the phenotypes of the offspring from this cross. The phenotypes will provide information on the recombinant and non-recombinant types.
- Determine Recombination Frequency: Calculate the recombination frequency by identifying the proportion of recombinant offspring compared to the total number of offspring. Recombination frequency is given by: Recombination Frequency = Number of Recombinant Offspring/Total Number of Offspring×100%
- Calculate Genetic Distance: The recombination frequency is often expressed in centimorgans (cM), where 1 cM corresponds to a 1% chance of recombination occurring between the two genes.
- Interpret the Results: If the recombination frequency is close to 50%, the genes are likely to be on different chromosomes or far apart on the same chromosome. A recombination frequency significantly less than 50% indicates that the genes are linked and located relatively close to each other on the same chromosome.
Advantages of Two-Point Cross
The two-point cross has several advantages in genetic mapping and analysis, including:
- Simplicity: The two-point cross is relatively straightforward to perform and analyze. It involves only two genes, making it easier to interpret the results compared to more complex crosses.
- Direct Measurement of Recombination Frequency: This method allows for the direct calculation of the recombination frequency between two genes, which is essential for determining their genetic distance on a chromosome.
- Identification of Linkage: A two-point cross can effectively identify whether two genes are linked (i.e., located close to each other on the same chromosome) or assort independently. This information is crucial for understanding inheritance patterns.
- Foundation for Further Mapping: The results from two-point crosses can serve as a foundation for more complex mapping techniques, such as three-point crosses, by providing initial estimates of gene distances and linkage relationships.
- Useful for Mapping New Genes: When mapping a new gene, a two-point cross can quickly provide information about its location relative to known genes, helping to establish its position on the genetic map.
- Cost-Effective: Since it involves fewer variables and simpler analysis, a two-point cross can be more cost-effective in terms of time and resources compared to more complex mapping strategies.
Limitations of Two-Point Cross
While two-point crosses are useful in genetic mapping, they also have several limitations:
- Underestimation of Genetic Distance: Two-point crosses can underestimate the actual genetic distance between genes, especially if the genes are far apart on the chromosome. This is because the method does not account for multiple crossover events that can occur between the genes.
- No Information on Gene Order: A two-point cross only provides information about the recombination frequency between two genes, but it does not reveal the order of multiple genes on a chromosome. This limitation makes it less informative when dealing with more than two genes.
- Limited to Two Genes: The two-point cross can only analyze the relationship between two genes at a time, which can be inefficient when studying multiple genes simultaneously. This limitation necessitates additional crosses to map more genes.
- Assumption of Independent Assortment: The two-point cross assumes that the genes being studied assort independently if they are located on different chromosomes. This assumption may not hold true for linked genes, leading to potential misinterpretation of results.
- Difficulty in Detecting Double Crossovers: Two-point crosses do not effectively detect double crossover events, which can lead to inaccuracies in estimating recombination frequencies and genetic distances.
- Potential for Confounding Factors: Environmental factors and genetic background can influence the results of a two-point cross, potentially confounding the interpretation of linkage and recombination frequencies.
Uses of Two-Point Cross
Two-point crosses are widely used in genetics for various purposes, including:
- Mapping Genetic Linkage: Two-point crosses are primarily used to determine whether two genes are linked (i.e., located on the same chromosome) and to estimate the distance between them based on recombination frequencies.
- Calculating Recombination Frequencies: By analyzing the offspring from a two-point cross, researchers can calculate the recombination frequency between two genes, which is essential for constructing genetic maps.
- Identifying Gene Interactions: Two-point crosses can help identify interactions between genes, such as epistasis, where the expression of one gene affects the expression of another. This information is crucial for understanding complex traits.
- Studying Inheritance Patterns: Two-point crosses can be used to study inheritance patterns in various organisms, helping to elucidate how traits are passed from one generation to the next.
- Testing Hypotheses: Researchers can use two-point crosses to test specific genetic hypotheses, such as the independence of gene assortment or the presence of linkage between genes.
- Mapping New Genes: When a new gene is discovered, a two-point cross can quickly provide information about its location relative to known genes, aiding in the construction of genetic maps.
- Breeding Programs: In agricultural and animal breeding programs, two-point crosses can be used to select for desirable traits by identifying linked genes that contribute to those traits.
- Genetic Counseling: Two-point crosses can assist in genetic counseling by providing information about the likelihood of inheriting specific traits or genetic disorders based on the linkage of genes.
- Understanding Genetic Variation: By analyzing recombination frequencies, two-point crosses can help researchers understand the genetic variation within populations and the evolutionary implications of that variation.
Example of Two-Point Cross
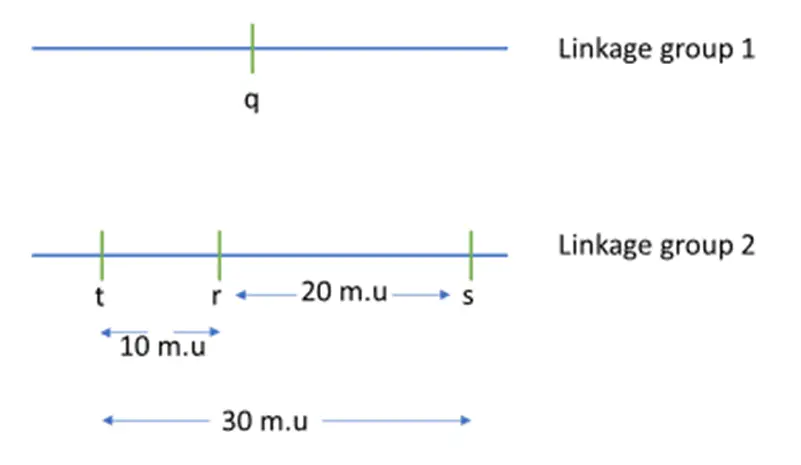
Recombination Frequencies for a Sequence of Two-Point Crosses for Four Genes (q, r, s & t)
| Gene loci | RF (%) |
|---|---|
| q and r | 50 |
| q and s | 50 |
| q and t | 50 |
| r and s | 20 |
| r and t | 10 |
Let’s break down the genetic mapping problem step by step based on the recombination frequencies (RF) provided for the four genes (q, r, s, and t):
Key Concepts
- Recombination Frequency (RF): The percentage of recombination between two genes, which helps in determining their relative distance on the chromosome. A higher RF suggests a greater distance or that the genes are on different chromosomes. An RF of 50% typically indicates independent assortment, meaning the genes are either on different chromosomes or very far apart on the same chromosome.
- Map Units (m.u.): The unit used to measure the distance between genes on a chromosome, where 1% RF equals 1 map unit.
Analysis
- Independent Assortment:
- RF = 50% for pairs (q, r), (q, s), and (q, t) indicates that genes q, r, s, and t are in different linkage groups or very far apart on the same chromosome. Therefore, q is not linked to r, s, or t, and these genes must be on different chromosomes or be in separate linkage groups.
- Linkage Analysis:
- RF between r and s is 20%, suggesting they are 20 map units apart.
- RF between r and t is 10%, suggesting they are 10 map units apart.
- Determining the Position of Gene t:
- To understand the position of gene t relative to gene r, compare the distance between r and s with the RF between s and t.
- If t is 10 m.u. to the left of r:
- The distance between t and s should be the sum of distances between r and s and between r and t.
- Expected distance between t and s would be approximately 30 m.u. (20 m.u. + 10 m.u.), but the RF between s and t is given as 28%, which suggests t is not 30 m.u. away but close to it.
- If t is 10 m.u. to the right of r:
- The distance between t and s should be approximately the difference between distances between r and s and r and t.
- Expected distance between t and s would be approximately 10 m.u. (20 m.u. – 10 m.u.).
- Conclusion:
- Since the RF between s and t is 28%, which is closer to the expected distance if t is to the right of r, it suggests that t is indeed to the right of r.
- Therefore, the distance between s and t should be about 10 m.u., fitting the given RF data.
Genetic Map
The genetic map based on the data would look like this:
- Order: r – t – s
- Gene q is not linked to the others and is on a different chromosome or linkage group.
What is Three-Point Cross?
A three-point cross is a genetic mapping technique that involves examining the recombination frequencies among three genes located on the same chromosome. This method is more informative than a two-point cross because it allows for the determination of the order of the three genes relative to each other, as well as the calculation of map distances between them.
In a three-point cross, a trihybrid individual (heterozygous for all three genes) is test-crossed with a homozygous recessive individual for those genes. The offspring produced from this cross are then analyzed to determine the frequency of parental and recombinant gametes. By comparing the frequencies of different offspring phenotypes, geneticists can identify which genes are linked and how far apart they are on the chromosome.
Methodology used in conducting a three-point cross
A three-point cross is a genetic cross used to determine the order and distance between three genes located on the same chromosome. Here’s a basic outline of the methodology:
- Select Parent Organisms: Choose two parent organisms that are homozygous for different alleles of the three genes you want to study. For example, if you’re studying genes A, B, and C, one parent might be homozygous for alleles A1, B1, and C1, and the other might be homozygous for alleles A2, B2, and C2.
- Create a Double-Heterozygous F1 Generation: Cross the two parent organisms to produce an F1 generation that is heterozygous for all three genes (A1A2, B1B2, C1C2).
- Cross F1 with a Double-Recessive Tester: The F1 individuals are then crossed with a tester organism that is homozygous recessive for all three genes (A1A1, B1B1, C1C1).
- Analyze the Offspring: The offspring from this cross will exhibit various combinations of the alleles. By analyzing the phenotypic ratios of these offspring, you can determine the recombination frequencies between the genes.
- Map the Genes: Use the recombination frequencies to determine the order and relative distances between the three genes. The gene with the highest recombination frequency from the other two is often located at the end of the chromosome, while the other two are closer together.
- Calculate Distances: The distances between genes are typically expressed in centimorgans (cM), where 1 cM corresponds to a 1% chance of recombination occurring between two genes.
Advantages of Three-Point Cross
The three-point cross offers several advantages in genetic mapping and analysis, including:
- Determination of Gene Order: One of the primary advantages of a three-point cross is its ability to determine the relative order of three genes on a chromosome. This is crucial for constructing accurate genetic maps.
- More Accurate Distance Measurements: The three-point cross allows for the calculation of genetic distances between genes while accounting for double crossover events. This leads to more accurate estimates of recombination frequencies compared to two-point crosses.
- Detection of Double Crossovers: This method can identify some double crossover events that may occur between the genes, which helps to correct for underestimations of genetic distance that can occur in two-point crosses.
- Efficiency in Mapping Multiple Genes: By analyzing three genes simultaneously, a three-point cross can provide more information in a single experiment compared to conducting multiple two-point crosses. This efficiency is particularly beneficial when mapping complex traits involving several genes.
- Improved Resolution: The three-point cross can provide better resolution in distinguishing between closely linked genes, which is important for understanding genetic interactions and inheritance patterns.
- Comprehensive Data: The data obtained from a three-point cross can be used to infer relationships among multiple genes, providing a more comprehensive understanding of genetic linkage and recombination.
- Foundation for Further Mapping: The results from three-point crosses can serve as a basis for more extensive mapping studies, allowing researchers to build upon existing knowledge of gene locations and relationships.
Limitations of Three-Point Cross
The three-point cross is a powerful tool in genetic mapping, but it also has several limitations:
- Complexity of Analysis: Analyzing the results of a three-point cross can be more complex than a two-point cross. The interpretation of recombination frequencies and the determination of gene order require careful consideration and can be prone to errors.
- Assumption of Linkage: The three-point cross assumes that the three genes being studied are linked. If any of the genes are unlinked or assort independently, the results may not accurately reflect their relationships, leading to incorrect conclusions.
- Underestimation of Distances: Similar to two-point crosses, three-point crosses can also underestimate genetic distances, particularly if double crossover events occur between the genes. While they can detect some double crossovers, not all may be accounted for, which can skew the distance calculations.
- Limited to Three Genes: The three-point cross can only analyze the relationships among three genes at a time. For mapping more than three genes, additional crosses are necessary, which can be time-consuming and resource-intensive.
- Potential for Confounding Factors: Environmental influences and genetic background can affect the outcomes of a three-point cross, potentially confounding the results and complicating the interpretation of linkage and recombination frequencies.
- Difficulty in Detecting Multiple Crossovers: While three-point crosses can identify some double crossover events, they may not effectively detect multiple crossover events involving more than two genes, which can lead to inaccuracies in mapping.
- Data Requirements: A three-point cross requires a sufficient number of offspring to provide reliable data for analysis. In cases where the number of progeny is low, the results may not be statistically significant.
Uses of Three-Point Cross
Three-point crosses are utilized in genetics for several important purposes, including:
- Determining Gene Order: One of the primary uses of a three-point cross is to establish the relative order of three genes on a chromosome. This is crucial for creating accurate genetic maps.
- Calculating Genetic Distances: Three-point crosses allow for the calculation of genetic distances between multiple genes while accounting for recombination events, including double crossovers. This leads to more precise estimates of linkage compared to two-point crosses.
- Detecting Double Crossovers: This method can identify some double crossover events that may occur between the genes, which helps to correct for underestimations of genetic distance that can occur in simpler mapping methods.
- Mapping Complex Traits: Three-point crosses are particularly useful for mapping complex traits that involve multiple genes. By analyzing three genes simultaneously, researchers can gain insights into the interactions and relationships among them.
- Improving Resolution of Genetic Maps: The three-point cross provides better resolution in distinguishing between closely linked genes, which is important for understanding genetic interactions and inheritance patterns.
- Facilitating Genetic Research: The data obtained from three-point crosses can be used to infer relationships among multiple genes, providing a more comprehensive understanding of genetic linkage and recombination.
- Foundation for Further Mapping Studies: The results from three-point crosses can serve as a basis for more extensive mapping studies, allowing researchers to build upon existing knowledge of gene locations and relationships.
- Enhancing Breeding Programs: In agricultural and animal breeding, three-point crosses can help identify and select for desirable traits by understanding the genetic basis of those traits and their linkage relationships.
- Understanding Genetic Mechanisms: By analyzing the outcomes of three-point crosses, researchers can gain insights into the mechanisms of genetic recombination and inheritance, contributing to the broader understanding of genetics.
How does a three-point cross improve upon the two-point cross method?
A three-point cross improves upon the two-point cross method in several significant ways:
- Increased Accuracy:
- The three-point cross allows for the detection of double crossover events, which can lead to underestimations of genetic distances in two-point crosses. By analyzing three genes simultaneously, researchers can more accurately calculate the distances between them.
- Determination of Gene Order:
- While two-point crosses can only provide information about the linkage between two genes, three-point crosses enable the determination of the relative order of three genes on a chromosome. This is crucial for constructing more detailed genetic maps.
- Comprehensive Data:
- A three-point cross provides more information by allowing the calculation of recombination frequencies between all pairs of genes (A-B, A-C, and B-C). This comprehensive data set helps in understanding the genetic relationships among multiple genes.
- Reduction of Ambiguity:
- The three-point cross reduces ambiguity in interpreting genetic linkage. In cases where two genes are closely linked, a two-point cross may not provide clear results. The three-point cross can clarify the relationships by providing additional context from the third gene.
- Better Mapping of Complex Traits:
- For traits influenced by multiple genes, three-point crosses are particularly useful. They allow researchers to map new mutations relative to previously mapped loci, facilitating the study of complex traits and their genetic basis.
- Enhanced Understanding of Genetic Interactions:
- By analyzing three genes at once, researchers can gain insights into how genes interact with one another, which is important for understanding epistasis and other genetic phenomena.
Define recombination frequency and its significance in genetic mapping.
Recombination Frequency is defined as the proportion of offspring in a genetic cross that exhibit a combination of traits different from those of the parents due to the exchange of genetic material during meiosis. It is calculated as the number of recombinant offspring divided by the total number of offspring, often expressed as a percentage or in map units (centimorgans, cM).
Significance in Genetic Mapping:
- Estimation of Genetic Distance:
- Recombination frequency is used to estimate the genetic distance between two genes on a chromosome. A higher recombination frequency indicates that the genes are farther apart, while a lower frequency suggests that they are closer together. This information is crucial for constructing genetic maps.
- Mapping Gene Locations:
- By determining the recombination frequencies between multiple pairs of genes, researchers can create a genetic map that shows the relative positions of genes on a chromosome. This helps in identifying the location of genes associated with specific traits or diseases.
- Understanding Linkage:
- Recombination frequency provides insights into whether genes are linked (located close together on the same chromosome) or assort independently (located on different chromosomes). Linked genes tend to have lower recombination frequencies.
- Identifying Genetic Interactions:
- Analyzing recombination frequencies can help in understanding genetic interactions and epistatic relationships between genes, which can influence phenotypic traits.
- Facilitating Breeding Programs:
- In agricultural and animal breeding, knowledge of recombination frequencies can assist in selecting for desirable traits by allowing breeders to track the inheritance of multiple genes simultaneously.
- Detecting Genetic Variability:
- Recombination contributes to genetic diversity within populations. By studying recombination frequencies, researchers can gain insights into the mechanisms of evolution and population genetics.
Compare and contrast two-point and three-point crosses in terms of accuracy and complexity.
Comparison of Two-Point and Three-Point Crosses
Accuracy
- Two-Point Crosses:
- Accuracy: Two-point crosses provide a basic estimate of the genetic distance between two genes based on recombination frequency. However, they can be less accurate due to the potential for underestimating distances caused by double crossover events, which may go undetected when only two genes are analyzed.
- Limitations: The accuracy of the distance estimation decreases as the distance between the two genes increases, particularly when recombination frequencies approach 50%, indicating independent assortment.
- Three-Point Crosses:
- Accuracy: Three-point crosses improve accuracy by allowing for the detection of double crossover events. This method provides a more comprehensive view of the genetic relationships among three genes, leading to more precise estimates of genetic distances.
- Gene Order Determination: Three-point crosses enable researchers to determine the order of genes on a chromosome, which enhances the accuracy of genetic mapping.
Complexity
- Two-Point Crosses:
- Complexity: Two-point crosses are relatively straightforward and simpler to perform. They involve analyzing the offspring from a cross between a heterozygous individual and a homozygous recessive individual for two genes, making the calculations and interpretations less complex.
- Data Analysis: The analysis focuses on a single pair of genes, which simplifies the data collection and interpretation process.
- Three-Point Crosses:
- Complexity: Three-point crosses are more complex as they involve analyzing three genes simultaneously. This requires a more intricate understanding of genetic interactions and the ability to calculate recombination frequencies for multiple pairs of genes (A-B, A-C, and B-C).
- Data Analysis: The analysis is more involved, as it includes accounting for double crossovers and determining the order of the genes. This complexity can lead to more comprehensive insights but requires more sophisticated data handling and interpretation.
What are the advantages of using three-point crosses over two-point crosses?
Using three-point crosses offers several advantages over two-point crosses in genetic mapping and analysis:
- Increased Accuracy:
- Three-point crosses allow for the detection of double crossover events, which can lead to underestimations of genetic distances in two-point crosses. This results in more accurate calculations of recombination frequencies and genetic distances between genes.
- Determination of Gene Order:
- A significant advantage of three-point crosses is the ability to determine the relative order of three genes on a chromosome. This is not possible with two-point crosses, which only provide information about the linkage between two genes.
- Comprehensive Data:
- Three-point crosses provide more information by allowing the calculation of recombination frequencies between all pairs of genes (A-B, A-C, and B-C). This comprehensive dataset helps in constructing more detailed and informative genetic maps.
- Reduction of Ambiguity:
- The three-point cross method reduces ambiguity in interpreting genetic linkage. In cases where two genes are closely linked, a two-point cross may not provide clear results. The three-point cross can clarify the relationships by providing additional context from the third gene.
- Better Mapping of Complex Traits:
- For traits influenced by multiple genes, three-point crosses are particularly useful. They allow researchers to map new mutations relative to previously mapped loci, facilitating the study of complex traits and their genetic basis.
- Enhanced Understanding of Genetic Interactions:
- By analyzing three genes at once, researchers can gain insights into how genes interact with one another, which is important for understanding epistasis and other genetic phenomena.
- Improved Efficiency in Mapping:
- Three-point crosses can be more efficient in mapping multiple genes simultaneously, reducing the number of crosses needed to establish gene order and distances compared to performing multiple two-point crosses.
- Facilitating Genetic Research:
- The ability to analyze three genes together can help in various genetic research applications, including the identification of genes associated with diseases, traits, and evolutionary studies.
How can three-point crosses be used to map complex traits in organisms?
Three-point crosses are a valuable method for mapping complex traits in organisms, following these steps and methodologies:
- Identifying Relevant Genes: Researchers start by pinpointing candidate genes believed to influence the complex trait of interest. This identification may stem from prior studies, known gene functions, or genetic markers linked to the trait.
- Creating a Trihybrid Organism: A trihybrid organism is generated by crossing individuals that are homozygous for different alleles at three loci (genes). The offspring will be heterozygous at all three loci, facilitating the analysis of recombination events among these genes.
- Performing a Test Cross: The trihybrid individual is then test-crossed with a homozygous recessive individual for all three genes. This cross yields offspring that can be examined for phenotypic traits, enabling researchers to observe the inheritance patterns of the alleles.
- Analyzing Offspring Phenotypes: Researchers record the phenotypes of the offspring, concentrating on the trait combinations resulting from allele recombination. By categorizing the offspring based on their phenotypes, they can distinguish between recombinant and parental types.
- Calculating Recombination Frequencies: Recombination frequencies between the three genes are determined by comparing the number of recombinant offspring to the total number of offspring. This data aids in estimating the genetic distances between the genes.
- Determining Gene Order: Utilizing the recombination frequencies, researchers can ascertain the order of the genes on the chromosome. This information is essential for understanding the arrangement of genes and their potential interactions in influencing the complex trait.
- Mapping Genetic Interactions: By examining the relationships among the genes, researchers can gain insights into how these genes interact to affect the complex trait. This includes identifying epistatic interactions, where the effect of one gene is modified by the presence of another.
- Identifying QTLs (Quantitative Trait Loci): Three-point crosses can assist in identifying QTLs associated with complex traits. By mapping the locations of genes contributing to trait variation, researchers can enhance their understanding of the trait’s genetic architecture.
- Facilitating Marker-Assisted Selection: In agricultural and breeding programs, the insights gained from three-point crosses can be utilized for marker-assisted selection. Breeders can select individuals with favorable allele combinations linked to desirable traits.
- Integrating Genomic Data: With advancements in genomic technologies, data from three-point crosses can be combined with genomic sequencing information to provide a more comprehensive understanding of the genetic basis of complex traits.
Describe a scenario in which a three-point cross would be preferred over a two-point cross.
Scenario: Mapping Flower Color, Plant Height, and Leaf Shape in a Plant Species
Background
A plant breeder aims to uncover the genetic factors influencing three specific traits: flower color (gene A), plant height (gene B), and leaf shape (gene C). Previous research suggests that these traits may be genetically linked, but the precise arrangement of the genes on the chromosome and their interactions remain unclear.
Step 1: Creating a Trihybrid
The breeder initiates the process by crossing two pure-breeding lines:
- One line features red flowers (A), tall plants (B), and round leaves (C).
- The other line has white flowers (a), short plants (b), and pointed leaves (c).
The resulting F1 generation will be heterozygous for all three traits (AaBbCc). The breeder then allows these F1 plants to self-fertilize, producing F2 offspring.
Step 2: Performing a Test Cross
To investigate the inheritance patterns, the breeder conducts a test cross by mating the F1 trihybrid (AaBbCc) with a triple homozygous recessive plant (aabbcc). This test cross generates offspring with various trait combinations, facilitating the observation of recombination events.
Step 3: Analyzing Offspring
The breeder collects data on the phenotypes of the F2 offspring, documenting the combinations of flower color, plant height, and leaf shape. For instance, the offspring may display:
- Red flowers, tall plants, round leaves (parental type)
- White flowers, short plants, pointed leaves (parental type)
- Recombinant types with different trait combinations.
Step 4: Calculating Recombination Frequencies
Using the phenotypic data, the breeder calculates the recombination frequencies between the three genes (A-B, A-C, and B-C). This information is vital for determining the genetic distances between the genes.
Step 5: Determining Gene Order
By analyzing the recombination frequencies, the breeder can ascertain the order of the genes on the chromosome. For example, if the recombination frequency between A and B is lower than that between A and C, and B and C, it indicates that the gene order is A-B-C.
Who developed the concepts of two-point and three-point crosses, and what was their impact on genetics?
The concepts of two-point and three-point crosses were developed by Stuart H. R. (Stuart H. R. H.) and George Beadle. Their work, particularly in the early to mid-20th century, was fundamental in the development of genetic mapping and chromosome theory.
Differences Between Two-Point and Three-Point Crosses
Two-point and three-point crosses are both methods used in genetic mapping, but they have distinct differences in their applications, methodologies, and the information they provide. Here are the key differences:
- Number of Genes Analyzed:
- Two-Point Cross: Involves the analysis of two genes at a time to determine their linkage and calculate the recombination frequency between them.
- Three-Point Cross: Involves the analysis of three genes simultaneously, allowing for the determination of gene order and the calculation of distances between all three genes.
- Information Provided:
- Two-Point Cross: Provides information about the linkage between two genes and the distance between them, but does not account for the arrangement of multiple genes.
- Three-Point Cross: Provides more comprehensive information, including the order of three genes on a chromosome and the distances between each pair of genes, while also accounting for double crossover events.
- Accuracy:
- Two-Point Cross: May lead to underestimations of genetic distances due to the inability to detect double crossover events.
- Three-Point Cross: Offers improved accuracy in measuring genetic distances by detecting some double crossover events, leading to more reliable genetic maps.
- Complexity:
- Two-Point Cross: Simpler in design and analysis, making it easier to perform and interpret.
- Three-Point Cross: More complex, requiring careful analysis of multiple gene interactions and recombination events, but yielding richer data.
- Applications:
- Two-Point Cross: Commonly used for initial mapping of genes and for studying simple inheritance patterns.
- Three-Point Cross: Used for more detailed genetic mapping, particularly when dealing with traits influenced by multiple genes, and for understanding gene interactions.
- Data Interpretation:
- Two-Point Cross: Results are typically presented as recombination frequencies between two genes.
- Three-Point Cross: Results include the order of genes and recombination frequencies among all three genes, allowing for a more comprehensive understanding of genetic relationships.
| Feature | Two-Point Cross | Three-Point Cross |
|---|---|---|
| Number of Genes Analyzed | Involves two genes | Involves three genes |
| Information Provided | Linkage and distance between two genes | Gene order and distances among three genes |
| Accuracy | May underestimate genetic distances | Improved accuracy by detecting double crossovers |
| Complexity | Simpler design and analysis | More complex, requiring detailed analysis |
| Applications | Initial gene mapping and simple inheritance studies | Detailed mapping of complex traits and gene interactions |
| Data Interpretation | Recombination frequencies between two genes | Order of genes and recombination frequencies among all three genes |
Fun Facts
- Did you know that Two-Point Crosses involve analyzing the recombination frequency between two genes to determine their relative distances on a chromosome, helping to construct simple genetic maps?
- Have you heard that Three-Point Crosses are used to map the positions of three genes simultaneously on the same chromosome, allowing scientists to determine gene order and distances more accurately?
- Are you aware that in Three-Point Crosses, the gene in the middle of the chromosome can be identified by looking at the recombinant classes that occur less frequently, which represent double crossovers?
- Did you know that the recombination frequency in Two-Point Crosses is often used to estimate the genetic distance between genes, where 1% recombination frequency equals 1 centimorgan (cM)?
- Can you believe that Three-Point Crosses can help identify double crossovers, which are recombination events that can complicate the interpretation of genetic distances and need to be corrected for accurate mapping?
- Have you ever heard that in a Three-Point Cross, the gene order can be determined by comparing the recombination frequencies and identifying the least frequent phenotypes as those resulting from double crossovers?
- Did you know that Two-Point Crosses provide information on linkage but can be limited in resolving the positions of multiple genes, while Three-Point Crosses offer a more detailed view of gene arrangements on a chromosome?
- Are you aware that the process of double crossovers can result in genetic distances between genes being underestimated when only pairwise recombination frequencies are considered?
- Did you know that Two-Point Crosses are typically used for simpler genetic analyses, while Three-Point Crosses are employed for more complex mapping studies involving multiple genes on the same chromosome?
- Can you believe that the data from Three-Point Crosses can reveal whether genes are located on the same chromosome and their relative positions, aiding in the construction of comprehensive genetic maps?
- https://opengenetics.pressbooks.tru.ca/chapter/mapping-with-three-point-crosses/#LongDesc11_3_4
- https://www.onlinebiologynotes.com/gene-mapping-two-point-test-cross-map-distance-and-frequency-of-recombination/
- https://passel2.unl.edu/view/lesson/18b30fa2ff29/4
- http://courseware.cutm.ac.in/wp-content/uploads/2020/05/Problems-on-two-point-test-cross.pdf