- Transfer RNAs, or tRNAs, are molecules that transport amino acids to the ribosome based on the nucleotide sequence of messenger RNA (mRNA). In this manner, they serve as the link between nucleotide and amino acid sequences.
- Because tRNAs are ribonucleic acids, they can establish hydrogen bonds with mRNA.
- In addition, they can create ester bonds with amino acids, allowing them to physically bring mRNA and amino acids together during the translation process.
- Each tRNA can base pair with a stretch of three nucleotides on mRNA. They pair with mRNA in a complementary and antiparallel way.
- These sequences of three nucleotides on the mRNA are known as codons, whereas the matching sequence on the tRNA is known as the anticodon.
- Base pairing between codon and anticodon imparts specificity to the translation process.
- A suitable amino acid is connected to the 3′ hydroxyl group of the tRNA based on the anticodon, and the ribosome catalyses the creation of a peptide bond between this amino acid and the elongating polypeptide chain.
What is tRNA?
- Transfer RNA, or tRNA, is a type of RNA that helps make proteins from mRNA.
- tRNA is used as an adapter molecule during the process of translation. It used to be called sRNA, which stands for soluble RNA.
- As an adapter, it connects the amino acids to the nucleic acids.
- It carries the amino acid that needs to be added to the peptide chain and also figures out the mRNA code for that amino acid.
- Most tRNAs have between 76 and 90 nucleotides. There are different tRNAs for each amino acid.
- No tRNA knows how to read stop codons.
- tRNA’s secondary structure looks like a clover leaf, and its tertiary structure looks like an upside-down “L.”
- The folded shape is caused by hydrogen bonds between bases that are different from each other.
History of tRNA
- Francis Crick’s “adaptor hypothesis” was the first theory about tRNA. This theory was based on the idea that there must be an adapter molecule that can translate the RNA alphabet into the protein alphabet.
- tRNA was found by Paul C. Zamecnik and Mahlon Hoagland. In the early 1960s, Alex Rich and Donald Caspar, two researchers from Boston, along with the Jacques Fresco group at Princeton University and a group from King’s College London in the United Kingdom, did a lot of important work on structure.
- Robert W. Holley of Cornell University wrote about the main structure and suggested three other structures in 1965. Robert M. Bock was the first person to find tRNA crystals. He did this in Madison, Wisconsin.
- In the years that followed, several other studies confirmed the cloverleaf shape[85], and in 1974, X-ray crystallography studies proved it for sure.
- Within a year, two different groups, one led by Alexander Rich and Kim Sung-Hou and the other by Aaron Klug in Britain, published the same crystallography results.
Properties of tRNA
- tRNAs are pieces of RNA that help turn mRNA into proteins.
- They have the shape of a cloverleaf, with a 3′ acceptor site, a 5′ terminal phosphate, a D arm, a T arm, and an anticodon arm.
- With the help of aminoacyl-tRNA synthetase, the main job of a tRNA is to carry amino acids to a ribosome complex on its 3′ acceptor site.
- Aminoacyl-tRNA synthetases are enzymes that put the right amino acid on a free tRNA so that proteins can be made.
- When an amino acid is attached to tRNA, it is called an aminoacyl-tRNA.
- The type of amino acid on a tRNA depends on the mRNA codon, which is a three-nucleotide sequence that tells the cell what amino acid to make.
- The anticodon is in the anticodon arm of the tRNA. It matches an mRNA codon and tells the tRNA which amino acid to carry.
- tRNAs can also control apoptosis by getting rid of cytochrome c.
tRNA biogenesis
- In eukaryotic cells, RNA polymerase III makes pre-tRNAs, which are the building blocks for tRNAs.
- Inside tRNA genes, RNA polymerase III recognises the 5′ intragenic control region (5′-ICR, D-control region, or A box) and the 3′ intragenic control region (T-control region, or B box).
- The first promoter starts at the +8 position of mature tRNAs, and the second promoter is 30–60 nucleotides after the first. When there are four or more thymidines in a row, the transcription stops.
- Inside the nucleus, pre-tRNAs go through a lot of changes. Some pre-tRNAs have introns that are cut out to make the functional tRNA molecule. In bacteria, these introns are removed by self-splicing, but in eukaryotes and archaea, they are removed by enzymes called tRNA-splicing endonucleases.
- Endonucleases need the bulge-helix-bulge (BHB) structure motif in eukaryotic pre-tRNA to find the tRNA intron and splice it correctly.
- The position and shape of this motif have stayed the same over time. But some organisms, like unicellular algae, have the 5′- and 3′-ends of the spliced intron sequence in a way that is not standard.
- The RNase P enzyme gets rid of the 5′ end, while the tRNase Z enzyme gets rid of the 3′ end.
- The archaeon Nanoarchaeum equitans is a notable exception because it doesn’t have an RNase P enzyme and its promoter is set up so that transcription starts at the 5′ end of the mature tRNA.
- A nucleotidyl transferase adds the 3′ CCA tail that doesn’t have a template.
- Before Los1/Xpo-t sends tRNAs into the cytoplasm, aminoacylation is done to the tRNAs.
- The processing events do not happen in the same order every time. For example, in yeast, splicing doesn’t happen in the nucleus but on the cytoplasmic side of the membranes that surround the mitochondria.
- Still, in March 2021, scientists reported evidence that an early form of transfer RNA may have been a replicator molecule in the very early stages of life’s development, a process called abiogenesis.
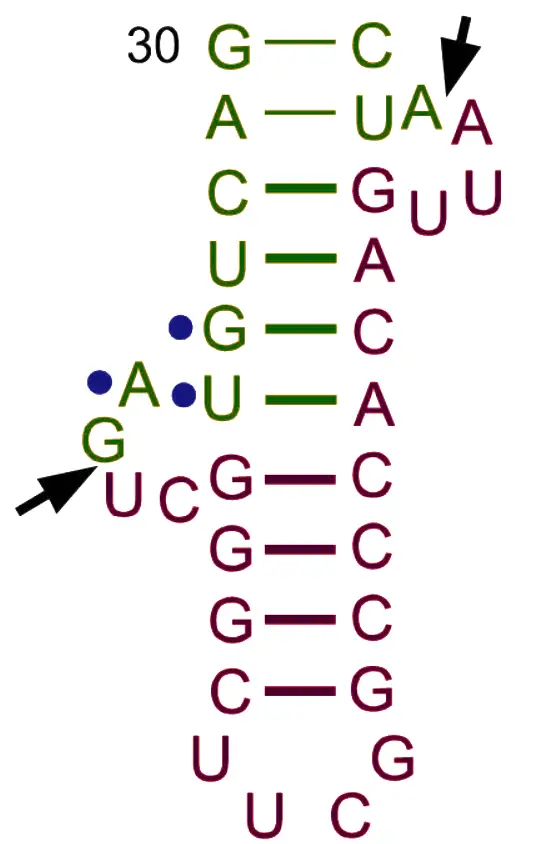
Structure of tRNA
The transfer RNA (soluble RNA) molecule has a molecular weight of about 25,000 and has between 71 and 80 nucleotides, most of which are 75. There are at least 20 different kinds of tRNAs, which is the same number of amino acids that are in proteins. Holley was the first person to figure out how tRNA (for alanine) is made.
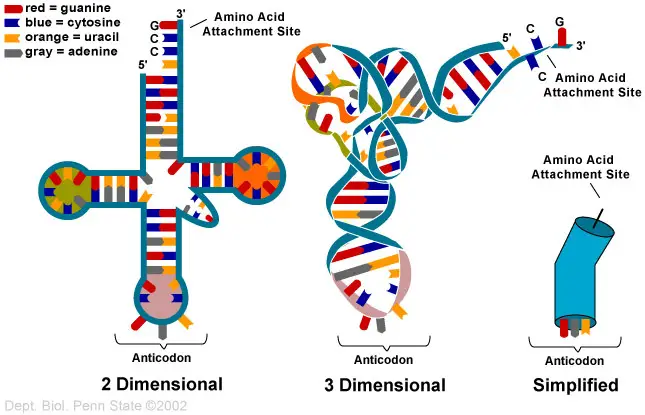
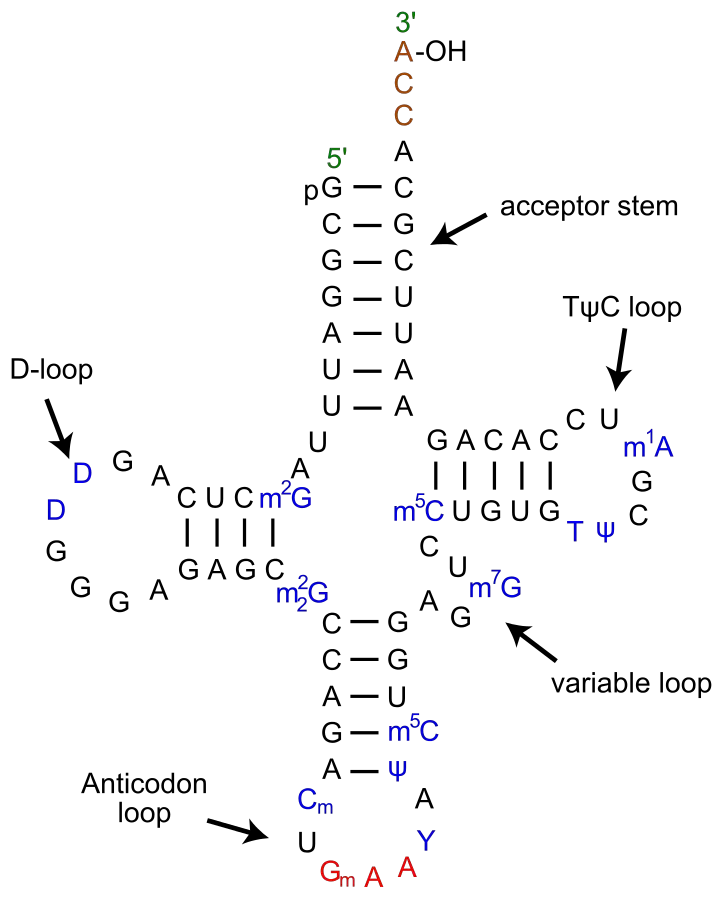
tRNA is made up of parts that look like a clover leaf. Most of the time, tRNA has four arms, and each arm has a base-paired stem.
Secondary Structure – Clover leaf structure of tRNA
The secondary structure of tRNA looks like a three-leafed clover because it has three hairpin loops. These are the main parts of tRNA:
1. Acceptor arm
- It is made when the bases of 7-9 nucleotides at the 5′ and 3′ ends pair up.
- The 3′ end has a specific sequence of CCA or CCA tails. The 5′ end has a phosphate group.
- The amino acid sticks to the acceptor arm’s 3′ hydroxyl group.
- The first step in the translation process is for tRNA to become charged with amino acids. The reaction is sped up by the enzyme aminoacyl tRNA synthetases.
2. DHU Loop
- The stem of the D arm has 3–4 base pairs, and it ends in a loop called the D loop because it usually has dihydrouridine, which is a modified nucleotide.
3. Anticodon Loop
- It has a long stem with 5 base pairs.
- It has a part called an anticodon loop that has the complementary codon (3 nucleotides) for the amino acid it carries. This is found on the mRNA.
- These bases in the anticodon loop that don’t have a partner pair with the mRNA codon.
- A different tRNA is used to find each codon.
4. TΨC Loop
- The T arm has a stem of 4-5 bp and a loop with pseudouridine, which is a changed form of uridine.
5. Variable Loop
- It is between the TΨC loop and the anticodon loop. It can be anywhere from 3 to 21 bases. It helps the tRNA molecule be recognised.
- This arm is the one that changes the most in tRNA. tRNAs are divided into two groups based on how different they are:
- Class I tRNAs: The most common form (about 75%), with a length of 3-5 base pairs.
- Class II tRNAs: They contain 13-20 base pair long arm.
Base pairs in tRNA:
The structure of tRNA is kept the same because the arms have complementary base pairs. Here are the four arms and the two pairs of their bases.
- The acceptor arm – 7 bp
- The TΨC arm – 5 bp
- The anticodon arm – 5 bp
- The D arm – 4 bp
Tertiary structure of tRNA
In 1968, crystals of tRNA were made, and x-ray crystallography was used to look at its 3-D structure. S.H.Kim came up with the most acceptable model (1973). He says that the 3-D shape of tRNA looks like the letter L. It is easy to get from the 2-D structure by combining the four arms of the clover leaf into two main domains –
- Acceptor arm – TΨC minihelix and
- Anticodon arm – D arm biloop.
The stem -CCA sticks out. Different tRNAs can also be told apart by comparing the angles of the two arms of the L-shaped structure. The corner of the L-shaped structure is made up of tertiary interactions between the TC- and D-loops. These interactions keep the structure stable.
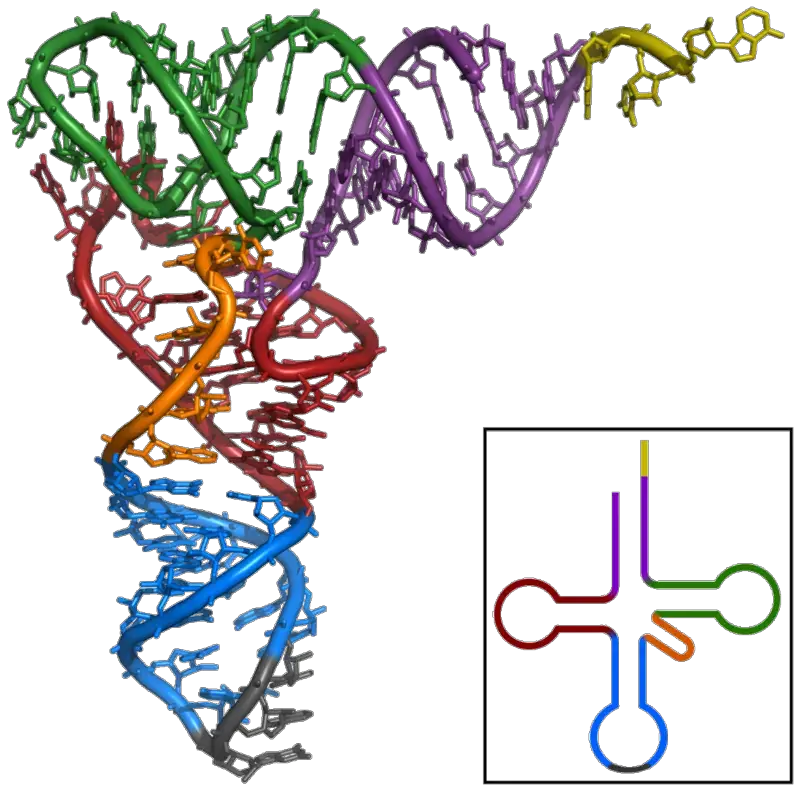
Properties of Tertiary structure
- It has a three-dimensional structure, and tRNA takes the shape of an L.
- This L-shape is made when the stem of the acceptor and the TC arms stack on one side, and the stem of the anticodon and the D arms stack on the other side. Both of these stacks make long helices, so they both fit together.
- Both of these long helices line up at a right angle, which also lines up the D-loop and the TC loop.
- The most stable structure is the tertiary structure, which is held together by hydrogen bonds between nitrogenous bases and between nitrogenous bases and the ribose-phosphate backbone.
- A tRNA molecule must have a tertiary structure so that it can read a codon in mRNA and make a protein.
Types of tRNA
- There are 20 different types of tRNA, which can be grouped by the amino acid they carry.
- They can also be put into groups based on their anticodon.
- When four nucleotides are put together, they can make 64 different codons. Three of these are stop codons, which tell the cell to stop translating.
- This means that one amino acid can be shown by more than one codon, and both the AATS and the tRNAs have to deal with this redundancy.
- But very few species have exactly 61 tRNAs. This makes me wonder how each tRNA knows which codon it needs to recognise.
- In many species, the number is much higher than 61, and different tRNAs with the same anticodon could have different translation efficiencies. This adds a layer of control to the process of making proteins.
- Through their anticodon loop, tRNAs talk to the codons on the mRNA. Base pairing between the codon and the anticodon makes sure that translation is specific.
- But the first base of the anticodon that matches the “wobble” or third position in a codon is usually changed so that the tRNA can form hydrogen bonds with three bases instead of just one.
- So, a single tRNA can recognise and pair with three different codons that all code for the same amino acid. Each of the 20 amino acids has its own AATS.
- This group of enzymes can find all the anticodons that stand for a certain amino acid. They are the second part of the machinery that handles redundant genetic code.
- Lastly, these molecules can be put into three groups: those with canonical amino acids attached to the right tRNA, those with wrongly attached amino acids, and those with modified amino acids like selenocysteine for non-canonical elongation.
Composition of tRNA
- A phosphate group at the 5′ end.
- The acceptor stem is a 7–9-base pair (bp) stem that is made by base pairing the 5′-terminal nucleotide with the 3′-terminal nucleotide, which has the CCA 3′-terminal group that is used to attach the amino acid. In general, these structures that look like tRNA at the 3′ end are called “genomic tags.” The acceptor stem may have base pairs that are not Watson-Crick pairs.
- The 3′ end of the tRNA molecule has a sequence of cytosine, cytosine, and adenine. This is called the CCA tail. The amino acid that aminoacyl tRNA synthetases add to the tRNA to make aminoacyl-tRNA is linked to the 3′-hydroxyl group on the CCA tail by a covalent bond. This sequence is important for enzymes to be able to find tRNA and is a key part of translation. In some tRNA sequences of prokaryotes, the CCA sequence is written down. The CCA sequence is added to most prokaryotic and eukaryotic tRNAs during processing and does not appear in the tRNA gene.
- The D arm is a stem with four to six base pairs that ends in a loop that often has dihydrouridine.
- The anticodon arm is made up of a 5-bp stem that has a loop that holds the anticodon. The anticodon is in the tRNA 5′-to-3′ primary structure, but it is in the wrong order because the mRNA needs to be read from 3′-to-5′.
- The T arm is a 4- to 5-bp stem that ends in a loop with the sequence TC, where is pseudouridine, which is a form of uridine that has been changed.
- Bases that have been changed, especially by methylation (by tRNA (guanine-N7-)-methyltransferase, for example), can be found in several places in the tRNA. The first anticodon base, or wobble-position, can be changed to inosine (from adenine), queuosine (from guanine), uridine-5-oxyacetic acid (from uracil), 5-methylaminomethyl-2-thiouridine (from uracil), or lysidine (derived from cytosine).
Magnesium as an important folding cofactor
- It is well known that magnesium ions are needed for many RNA systems to work properly. This is true for tRNAs as well.
- In line with crystallographic data, NMR experiments show that four strong binding sites are clustered in the D stem and its junction with the acceptor stem and the anticodon stem, and that tRNAs can take on different shapes when magnesium ions are not present.
- So, magnesium cations make tRNA structures more rigid, and it is generally agreed that a tRNA molecule needs up to four magnesium cations to be stable.
- It has also been shown that when E. coli tRNAGlu is made without Mg2+, it forms a stable conformer that is inactive in the aminoacylation reaction.
- In other words, magnesium is needed for this tRNA to change into a shape that can be aminoacylated.
- Without a doubt, magnesium cations, along with strongly bound monovalent cations and water molecules, are important parts of tRNA structures and are needed to make active RNA folds and keep them stable.
What do you mean by Post-Transcriptional Modification of tRNA?
- In the human genome, there are almost 500 genes that code for tRNAs and 300 gene fragments that are linked to these RNAs.
- RNA polymerase III copies these genes, and the copied RNA goes through a lot of changes, especially in eukaryotes.
- Endonucleases work on the boundary between an intron and an exon, the 5′ and 3′ ends of the RNA are changed, and enzymes add the last CCA residues to the 3′ end of the tRNA.
- The aminoacylation of the CCA residues could happen in the nucleus, and the charged tRNA could then leave the nucleus.
- Also, many of the bases on the tRNA are changed, especially through methylation (adding a methyl group) and deamidation (removal of an amide group).
- In particular, the first base of the anticodon that pairs with the “wobble” position on the codon is changed to allow unusual types of base pairing.
- Adenine can be changed into inosine, which makes it possible for uracil, cytosine, and adenine to pair with it.
- Pseudouridine is an enzyme-driven isomerization of uridine residues that makes it a common modified base. It is thought to contribute to the stability of the tRNA molecule by making the nearby sugar-phosphate backbone more rigid and by affecting how the bases in nearby regions are stacked.
- Lysidine is an unusual base that is made when a cytidine residue is joined to a lysine amino acid.
- The isoleucine tRNA uses the fact that lysidine only pairs with adenosine to make sure that the translation is specific.
- Based on the anticodon, AATS attaches the right amino acid to the tRNA molecule. These enzymes have places where they can bind to the amino acid, tRNA, and ATP. They break down ATP into AMP and attach the amino acid to the ribose sugar of the last nucleotide on tRNA.
- The tRNA is now considered to be “charged” and can take part in the reactions on the ribosome that make proteins.
- This reaction usually happens in the cytoplasm, but it has been seen in the nucleus as well.
- The enzyme binds to many parts of the tRNA to make sure that the reaction is very specific. It even checks its own reaction for mistakes because the structures of many amino acids are similar.
- The RanGTP system is then used to send the mature tRNA out of the nucleus by binding it to certain export factors.
- In this process, the acceptor arm and T-arm are very important. There is also a lot of interaction between the export factors and the RNA molecule, which means that only fully processed and complete tRNAs can move to the cytoplasm.
How does tRNA Interact with ribosomes?
- The ribosome has three important parts: the P (peptidyl) site, which holds the growing polypeptide, the A (acceptor) site, which takes in a new charged tRNA, and the E (exit) site, through which a deacylated tRNA leaves the ribosome.
- These places are on both parts of the ribosome. They are called P/P or A/A sites, depending on which part of the ribosome they are on.
- For example, the P/P site binds to tRNA that is anchoring a polypeptide chain, and the A/A site is anchoring a charged tRNA that is coming in.
- The growing polypeptide is passed from the peptidyl-tRNA at the P/P site to the tRNA at the A/A site, where it is broken down. To keep translating, the ribosome moves forward by one codon. This makes the tRNA at the P/P site switch to a temporary P/E configuration and then to the E/E configuration before leaving the ribosome.
- In the same way, the tRNA at the A/A site takes on a temporary A/P binding shape before settling at the P/P site, ready for the next amino acid to continue translation.
What is Anticodon?
- An anticodon is a group of three nucleotides that match the three bases of an mRNA codon.
- Each tRNA has a unique anticodon triplet sequence that can pair up with one or more codons for an amino acid to make three complementary base pairs.
- Due to wobble base pairing, some anticodons pair with more than one codon. Often, the first nucleotide of the anticodon is not found on the mRNA. This nucleotide is inosine, which can form hydrogen bonds with more than one base in the position of the corresponding codon.
- In the genetic code, it is common for a single amino acid to be specified by all four third-position possibilities, or at least by both pyrimidines and purines. For example, the amino acid glycine is coded for by the sequences GGU, GGC, GGA, and GGG. At the first anticodon position, which is sometimes called the “wobble position,” other modified nucleotides can also show up. This makes small changes to the genetic code, like in mitochondria.
- As there are 61 sense codons in the standard genetic code, each cell needs 61 different types of tRNA to match up with the 61 sense codons that specify amino acids.
- But many cells have less than 61 types of tRNAs because the wobble base can bind to some, but not all, of the codons that tell the cell to make a certain amino acid.
- At least 31 tRNAs are needed to be able to translate all 61 sense codons in a clear way.
What is Aminoacylation?
- Adding an aminoacyl group to a compound is the process of aminoacylation.
- It bonds an amino acid to the CCA 3′ end of a tRNA molecule in a way that can’t be broken down.
- An aminoacyl tRNA synthetase adds a certain amino acid to each tRNA. This is called “charging” the tRNA.
- Even though there can be more than one tRNA and more than one anticodon for an amino acid, there is usually only one aminoacyl tRNA synthetase for each amino acid.
- Synthetases don’t just use the anticodon to find the right tRNA; the acceptor stem is often a very important part of the process. Reaction:
- amino acid + ATP → aminoacyl-AMP + PPi
- aminoacyl-AMP + tRNA → aminoacyl-tRNA + AMP
- Some living things can be missing one or more aminophosphate-tRNA synthetases. This causes a chemically related amino acid to charge the tRNA. Then, an enzyme or enzymes change the tRNA so that it has the right charge.
- Helicobacter pylori, for example, does not have glutaminyl tRNA synthetase.
- So, glutamate tRNA synthetase adds glutamate to the end of tRNA-glutamine (tRNA-Gln).
- The acid side chain of the glutamate is then changed into an amide by an enzyme called an amidotransferase. This makes the correctly charged gln-tRNA-Gln.
- Interfering with aminoacylation could be a way to treat some diseases. For example, cancer cells may be more sensitive to changes in aminoacylation than healthy cells.
- Cancer and viral biology are often very dependent on certain tRNA molecules when it comes to making proteins.
- For example, feeding tRNA-Lys-CUU with lysine keeps liver cancer cells growing and spreading, while healthy cells depend much less on this tRNA for their normal function.
- In a similar way, the hepatitis E virus needs a tRNA landscape that is very different from that of cells that are not infected.
- So, stopping the aminoacylation of certain tRNA species is seen as a promising new way to treat a wide range of diseases in a logical way.
Functions of tRNA
- tRNA is an important part of the process of making proteins.
- It connects amino acids to their specific codons in mRNA by acting as an adapter molecule.
- The first step in making a protein is for tRNA to become aminoacylated.
- tRNA knows which amino acid it needs to carry and does so in the ribosomal subunits as part of the translation process.
- The tRNA moves the amino acid to the growing polypeptide chain in the ribosome, which has three binding sites for tRNA: A, P, and E, which stand for aminoacyl, peptidyl, and exit, respectively.
- Codons on mRNA are decoded by certain tRNAs until the whole sequence of a polypeptide chain has been translated.
References
- Schultz, D. W., & Yarus, M. (1994). tRNA Structure and Ribosomal Function. Journal of Molecular Biology, 235(5), 1381–1394. doi:10.1006/jmbi.1994.1095
- Lorenz C, Lünse CE, Mörl M. tRNA Modifications: Impact on Structure and Thermal Adaptation. Biomolecules. 2017; 7(2):35. https://doi.org/10.3390/biom7020035
- Westhof, E., & Auffinger, P. (2012). Transfer RNA Structure. eLS. doi:10.1002/9780470015902.a00005
- https://ib.bioninja.com.au/higher-level/topic-7-nucleic-acids/73-translation/ribosomes-and-trna.html
- http://www.lscollege.ac.in/sites/default/files/e-content/tRNA%20Structure.pdf
- http://www.lscollege.ac.in/sites/default/files/e-content/tRNA%20Structure.pdf
- https://www.vedantu.com/biology/trna
- https://www.slideshare.net/HarshitaBhawsar/trna-structure-and-function-60453762
- http://www2.tulane.edu/~biochem/nolan/lectures/rna/trnaz.htm
- https://www.nature.com/scitable/definition/trna-transfer-rna-256/
- https://www.ncbi.nlm.nih.gov/books/NBK6236/
- https://thebiologynotes.com/transfer-rna/
- https://earth.callutheran.edu/Academic_Programs/Departments/BioDev/omm/jsmolnew/trna/trna.html
- https://www.khanacademy.org/science/biology/gene-expression-central-dogma/translation-polypeptides/a/trna-and-ribosomes
- https://en.wikipedia.org/wiki/Transfer_RNA
- https://pdb101.rcsb.org/motm/15
- https://www.ias.ac.in/article/fulltext/jbsc/031/04/0453-0457
- https://biologydictionary.net/trna/
- https://www.sciencedirect.com/science/article/abs/pii/S0022283684710953