Torovirus is a genus of enveloped, positive-strand RNA viruses belonging to the Nidovirales order and Tobaniviridae family.They infect predominantly vertebrates, particularly cattle, pigs, and horses. Among the diseases associated with this genus is gastroenteritis, a condition typically observed in mammals.Torovirus is the only genus in the monotypic Torovirinae subfamily.Torovirus is a monotypic taxon with a single subgenus, Renitovirus.
The initial discovery of the torovirus dates back to the 1970s. Equine torovirus (EToV) was discovered by accident in the rectal sample of a horse with severe diarrhea. In 1979, the ‘Breda’ bovine torovirus was discovered during an investigation at a Breda dairy farm. Several heifers suffered from severe diarrhea for months. In 1984, the electron microscope (EM) was used to detect torovirus-like particles in patients with gastroenteritis.
Torovirus
Equine torovirus (EToV), formerly known as Berne virus, was inadvertently isolated in 1972 in equine kidney cells using a rectal swab from a horse with diarrhea. EToV is the only torovirus that has been propagated in cell culture, in equine dermis or embryonic mule skin cell lines, where it causes a cytopathic effect that results in cell death. While Berne virus was not neutralized by antisera against known equine viruses, serologic cross-reactions were observed in neutralization tests and enzyme-linked immunosorbent assays (ELISA) using sera from calves experimentally infected with morphologically similar particles, then known as ‘Breda’ viruses.
In 1979, during an investigation of a dairy herd in Breda, Iowa, where acute neonatal calf diarrhea had been a problem for three consecutive years, the first Breda virus, now known as bovine torovirus (BToV), was discovered. After this initial report, additional BToV strains were identified in cattle calves from Ohio and a colostrum-deprived (CD) calf from Iowa. BToV cannot be adapted to grow in cell or tissue cultures and must be passed in gnotobiotic (Gn) calves, impeding its biochemical, biophysical, and molecular characterization despite repeated attempts. The majority of studies on the pathogenesis and pathology of torovirus infections have been conducted on BToV-infected Gn and CD heifers, as well as in limited field studies; in contrast, the majority of data on the biochemistry and morphogenesis of toroviruses has been derived from EToV studies.
Using electron microscopy (EM), torovirus-like (TVL) particles were detected in the feces of patients with gastroenteritis in 1984. Several countries have since received reports of human toroviruses (HToVs) in infants and adults with acute diarrhea. TVL particles have also been detected in the feces of swine, where they have been designated as porcine torovirus (PToV). The discovery of toroviral RNA sequences in the excrement of piglets and diarrhea-afflicted humans provided evidence that the observed structures were not artifacts. In recent years, TVL particles have also been found in poultry and have been linked to a’stunting syndrome’.
Classification of Torovirus/Taxonomy and Classification
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Orthornavirae |
| Phylum: | Pisuviricota |
| Class: | Pisoniviricetes |
| Order: | Nidovirales |
| Family: | Tobaniviridae |
| Subfamily: | Torovirinae |
| Genus: | Torovirus |
Toroviruses are positive-polarity, single-stranded RNA viruses with an envelope containing peplomers. It was the unique biconcave disk and C-shape of the virion in the extracellular environment that led to this nomenclature. Based on similarities in genomic organization and replication strategies, the genus Torovirus has been included with the genus Coronavirus and the newly recognized genus Bafinivirus in the family Coronaviridae since 1992. Additionally, toro- and coronaviruses share a common ancestor from which their polymerase and envelope genes diverged. Toroviruses have adopted the nomenclature for coronavirus genes, mRNAs, and structural proteins due to their membership in the family Coronaviridae. However, the absence of sequence homology in structural genes and antigenic relatedness with coronaviruses justifies their taxonomic classification as a distinct genus. Currently, the International Committee on Virus Taxonomy (ICTV) recognizes four species within the genus Torovirus: equine torovirus, bovine torovirus, porcine torovirus, and human torovirus.
The families Coronaviridae, Arteriviridae, and the newly described family Roniviridae comprise the order Nidovirales, the second order in animal virology (following the order Mononegavirales). This assignment is based on their similar basic genomic organization and shared replication strategy: the synthesis of a 3′ co-terminal nested set of subgenomic mRNAs and the presence of two open reading frames (ORFs) connected by a frameshift site in order to express a replicase directly from the genomic RNA. This nested set of mRNAs served as the basis for the naming of the order Nidovirales (from Latin nidus, meaning “nest”).
Structure of Torovirus
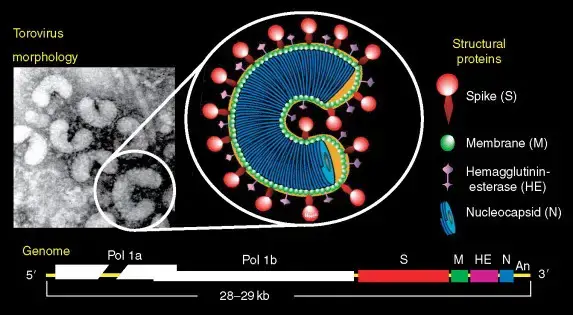
- Torovirus particles have a nucleocapsid with helical symmetry that is wound into a hollow tube (diameter 23 nm, average length 104 nm, and periodicity 4.5 nm).
- Torovirus virions are typically observed as kidney- or C-shaped particles (105–140 nm × 12–20 nm) that are negatively stained extracellularly. Depending on the orientation of the virions with respect to the electron beam, they can also be observed as spherical or oval particles (89 ± 7 nm × 75 ± 9 nm) or rod-shaped virions (35 nm × 170 nm).
- A 11-nm-thick envelope envelops the virion structure, which is characterized by conspicuous drumstick-shaped peplomers (17–24 nm) and a fringe of shorter spikes (8–10 nm) that represent the spike and hemagglutinin-esterase (HE) proteins, respectively.
- In the cytoplasm of infected intestinal cells, toroviruses are observed as elongated tubules with rounded extremities (rod-shaped virions, 35–42 nm 80–105 nm).
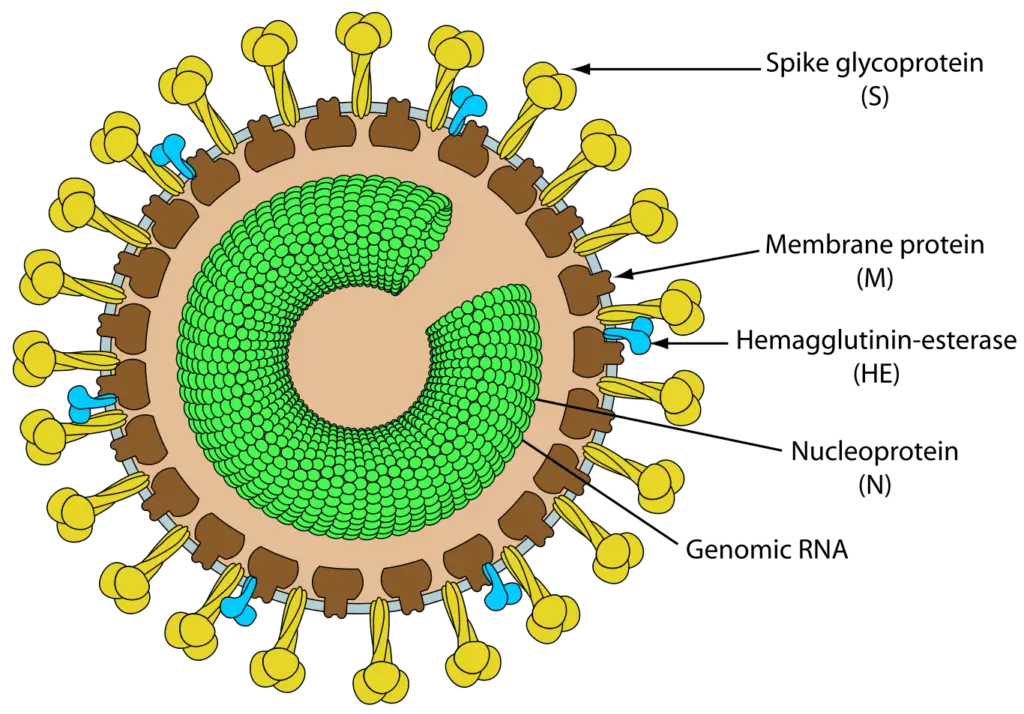
Toroviruses (ToV) are single-stranded RNA viruses with an envelope containing peplomers that are frequently associated with enteric infections in cattle and possibly humans. ToVs have been reported in Europe, the Americas, New Zealand, and South Africa, as well as countries on other continents.Torovirus particles typically have a nucleocapsid that is coiled into a hollow cylinder shape. The diameter is approximately 23 nm and the average length is 104 nm, with 4.5 nm between each turn cycle.ToVs are pleomorphic, spanning in size from 100 to 150 nm, with club-like protrusions emanating from the capsid. Toroviruses also possess a nucleocapsid with helical symmetry and a donut-shaped nucleocapsid. Equine torovirus (EToV) is the only species of Torovirus that can be grown in a cell culture medium, so it has been the subject of the most research. Immunofluorescence investigations and the morphology of the intestinal cells of bovine Toroviruses (BToV) have revealed similarities between EToVs and BToVs. Torovirus and members of the related family Coronaviridae are both round, pleomorphic, enveloped viruses measuring between 120 and 140 nm in diameter.
Genome of Torovirus
- The torovirus genome consists of a single-stranded, polyadenylated RNA with positive (messenger) polarity and a length of approximately 28.5 kbp.
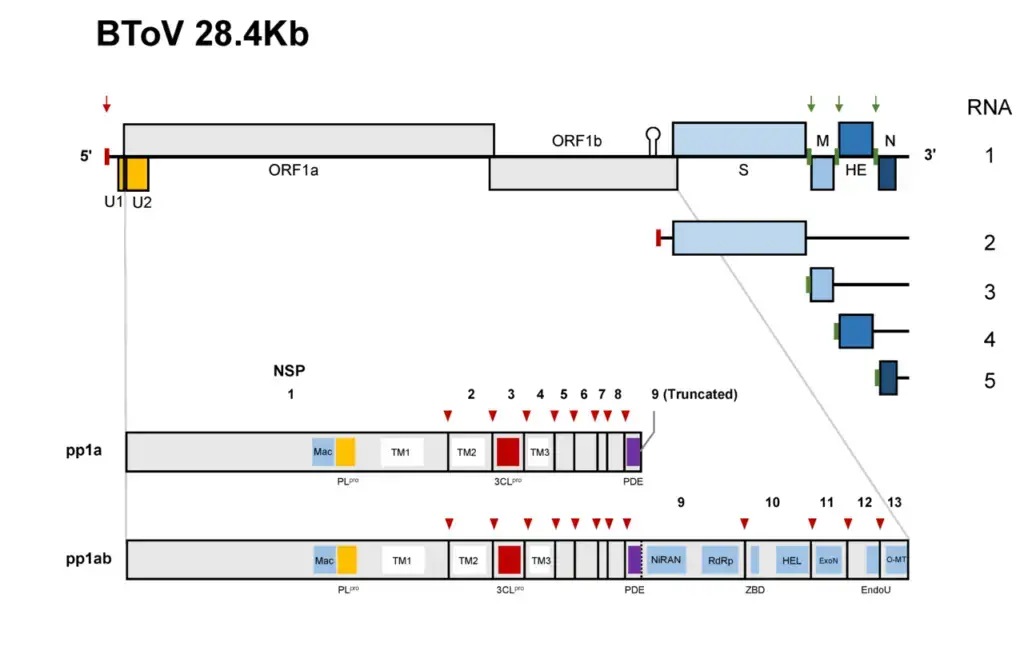
- Recent genome sequencing has revealed that the BToV genome contains 28,475 nucleotides and six ORFs, each of which encodes a known protein. ORF1a and ORF1b comprise the replicase (RNA-dependent RNA polymerase) gene, which is expressed as a large precursor protein directly from the genomic RNA via a ribosomal frameshift mechanism, similar to other nidoviruses.
- Apparently, the large product of these ORFs is implicated in the synthesis of negative-strand RNA and the initiation of genomic and subgenomic RNA synthesis.
- The remaining four ORFs encode structural protein genes and are expressed through the production of a 3′ co-terminal nested set of four mRNAs.
- The BToV ORF2 (4752 nucleotides), ORF3 (702 nucleotides), ORF4 (1248–1251 nucleotides), and ORF5 (504 nucleotides) genes code for the spike (S), membrane (M), HE, and nucleocapsid (N) proteins, respectively.
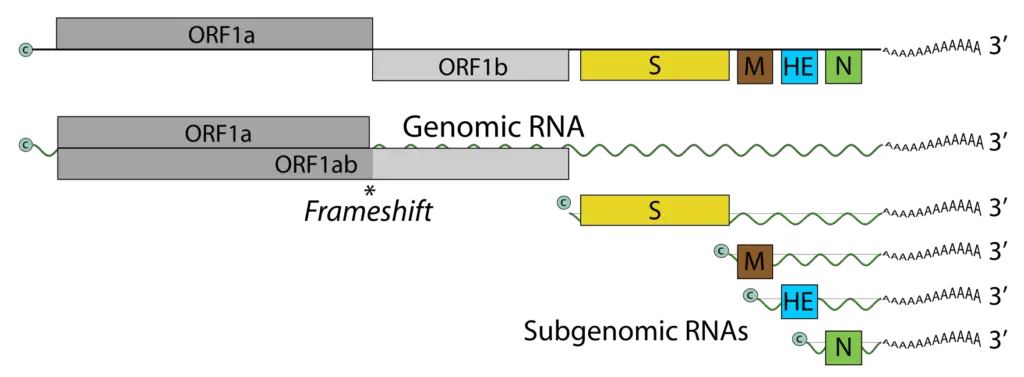
The infectious virion RNA functions as both the viral genome and viral messenger RNA. As with ORF1b, ORF1a is translated via ribosomal frameshifting from RNA encoding. RNA synthesis involves the processing of pp1a and pp1b into the viral polymerase (RdRp) and other non-structural proteins. Subgenomic RNAs are expressed as structural proteins. Each genomic and subgenomic RNA is translated to produce only the protein encoded by the 5′-most open reading frame (ORF). A hypothetical 30K protein that overlaps ORF1a may be expressed from an alternate CUG codon.
Proteins of Torovirus
- In EToV virions, proteins with molecular weights of 20, 22, 37, and 80–100 kDa have been identified. The release of the 22, 37, and 80–100 kDa proteins upon detergent treatment of virions indicates their association with the envelope.
- Using surface radioiodination, similar polypeptides of 20, 37, 85, and 105 kDa have been identified in purified BToV. With a predicted molecular weight of 18.3–19.0 kDa, the nucleocapsid (N) protein (167 amino acids) is the most abundant polypeptide in the virion (80–84%).
- Blotting experiments with EToV have revealed that this internal protein is the only RNA-binding polypeptide in the virion. M glycoprotein (233 amino acids, 26.5 kilodaltons) is the second most prevalent protein (∼13%) and is likely associated with the envelope.
- Three consecutive transmembrane -helices are found in the N-terminal half of a class III membrane protein that lacks a signal sequence but contains three consecutive transmembrane -helices.
- During the budding process, the M protein accumulates in intracellular membranes, primarily those of the endoplasmic reticulum, and is thought to perform a role in assembly, maturation, and nucleocapsid recognition.
- The heterogeneous, N-glycosylated, 80–100 kDa protein is recognized by both neutralizing and hemagglutination-inhibiting monoclonal antibodies, thus identifying it as the spike (S) protein protruding from the virion surface.
- The spike (S) gene encodes an apoprotein (1581 amino acids) of approximately 178 kDa. The predicted amino acid sequence contains domains characteristic of type I membrane glycoproteins, including an N-terminal signal sequence, a putative C-terminal transmembrane anchor, and a cytoplasmic tail.
- The HE is a class I membrane N-glycosylated protein (416–418 amino acids) with a molecular weight of 65 kDa that is also found in the BToV envelope. It possesses an N-terminal signal sequence, a C-terminal transmembrane domain, a number of N-glycosylation sites, and a putative ‘FGDS’ motif with acetylesterase activity specific for N-acetyl-9-O-acetylneuraminic acid.
- The HE protein may be a second receptor-binding protein (in addition to the spike protein), but it is not involved in viral entry. EToV virions lack HE protein and possess only a partial ORF4 sequence.
Physical Properties of Torovirus
- In EToV virions, proteins with molecular weights of 20, 22, 37, and 80–100 kDa have been identified. The release of the 22, 37, and 80–100 kDa proteins upon detergent treatment of virions indicates their association with the envelope.
- Using surface radioiodination, similar polypeptides of 20, 37, 85, and 105 kDa have been identified in purified BToV. With a predicted molecular weight of 18.3–19.0 kDa, the nucleocapsid (N) protein (167 amino acids) is the most abundant polypeptide in the virion (80–84%).
- Blotting experiments with EToV have revealed that this internal protein is the only RNA-binding polypeptide in the virion.
- M glycoprotein (233 amino acids, 26.5 kilodaltons) is the second most prevalent protein (13%) and is likely associated with the envelope. Three consecutive transmembrane -helices are found in the N-terminal half of a class III membrane protein that lacks a signal sequence but contains three consecutive transmembrane -helices.
- During the budding process, the M protein accumulates in intracellular membranes, primarily those of the endoplasmic reticulum, and is thought to perform a role in assembly, maturation, and nucleocapsid recognition.
- The heterogeneous, N-glycosylated, 80–100 kDa protein is recognized by both neutralizing and hemagglutination-inhibiting monoclonal antibodies, thus identifying it as the spike (S) protein protruding from the virion surface.
- The spike (S) gene encodes an apoprotein (1581 amino acids) of approximately 178 kDa. The predicted amino acid sequence contains domains characteristic of type I membrane glycoproteins, including an N-terminal signal sequence, a putative C-terminal transmembrane anchor, and a cytoplasmic tail.
- The HE is a class I membrane N-glycosylated protein (416–418 amino acids) with a molecular weight of 65 kDa that is also found in the BToV envelope. It possesses an N-terminal signal sequence, a C-terminal transmembrane domain, a number of N-glycosylation sites, and a putative ‘FGDS’ motif with acetylesterase activity specific for N-acetyl-9-O-acetylneuraminic acid.
- The HE protein may be a second receptor-binding protein (in addition to the spike protein), but it is not involved in viral entry. EToV virions lack HE protein and possess only a partial ORF4 sequence.
Replication and Morphogenesis of Torovirus
EToV morphogenesis has been the most thoroughly studied because it can be grown in cell culture. Immunofluorescence (IF) and ultrastructural examinations of BToV-infected calf intestinal cells have revealed similarities with EToV morphogenesis.
Attachment, Entry, and Uncoating
- Attachment to the apical surface of enterocytes appears to be mediated by spike proteins, but HE proteins may also be involved. BToV appears to infiltrate or penetrate enterocytes via receptor-mediated endocytosis.
- Uncoating and subsequent release of BToV RNA is most likely caused by lysosomal degradation in virus-containing vesicles. The location of this phenomenon has not yet been determined.
- The EToV replicates in the cytoplasm. Preformed tubular capsids proliferate through the Golgi stack and endoplasmic reticulum membranes. UV (ultraviolet) pre-irradiation of cells, actinomycin D, and α-amanitin are reported to reduce virus yields, indicating that a nuclear function of the host cell is required. The duration of the replication cycle is approximately 10–12 hours.
Transcription
- In EToV-infected cells, five polyadenylated mRNAs with sizes of >20.0, 7.5, 2.1, 1.4, and 0.8 kbp are detected.
- The five EToV mRNAs form a 3′ co-terminal nested set, as revealed by Northern (RNA) blot hybridizations with restriction fragments from cDNA clones. Sequence analysis reveals the presence of four complete ORFs whose initiation codons coincide with the 5′ extremities of EToV RNAs 2–5; RNA 5 is contiguous to the consensus sequence.
- UV transcription mapping has established that EToV RNAs 1–3 are independently transcribed. The genes for M, HE, and N are preceded by brief noncoding ‘intergenic’ regions that contain a transcription-regulating element (TRE) with the consensus sequence 5′ (C)ACN3–4CUUUAGA 3′.
- A duplicate of this sequence exists at the extreme 5′ end of the genome. In contrast, the S gene coincides with the replicase gene and the N-terminal region of the replicase gene.
- In fact, 28 S residues are encoded by an internal (–1) reading frame within ORF1b, and there is no TRE. This 3′ co-terminal nested set of mRNAs justifies the incorporation of toroviruses within the order Nidovirales.
Translation
- There is no RNA-dependent RNA polymerase present in torovirus particles. Torovirus replicase is most likely translated as soon as RNA is released.
- Translation generates two large polyproteins, from which proteolytic cleavage derives the various subunits of the viral replicase/transcriptase as well as accessory proteins with as-yet-unidentified functions.
- The genes for the structural proteins S, M, HE, and N are located 5′ to 3′ upstream of ORF1b; they are translated from four subgenomic mRNAs enumerated 2–5 (with the genomic RNA as RNA1).
Post-Translational Processing
- The N-glycosylated S protein results from the metabolism of a 200 kDa precursor that is present in infected cells but not in virions.
- The amino acid sequence of the spike protein contains eighteen potential N-glycosylation sites, two heptad repeat domains, and a putative ‘trypsin-like’ cleavage site.
- The mature S protein is composed of two subunits, and the electrophoretic mobility of these subunits upon treatment with endoglycosidase F suggests that the predicted cleavage site is functional in vivo.
- The heptad repeat domains are likely involved in the formation of an intrachain coiled-coil secondary structure; similar interchain interactions may contribute to the observed formation of S protein dimers. The intrachain and interchain coiled-coil interactions may stabilize the torovirus peplomer stalk.
Assembly, Budding, Egress, and Maturation
- Approximately 10 hours after infection, EToV particles are detected both intracellularly and extracellularly.
- Tubular structures of variable length, diameter, and electron density appear in the cytoplasm and nucleus of infected cells at this time, likely representing nucleocapsids in their preformed state. It is unknown whether the accumulation of nucleocapsids in the nucleus reflects a nuclear phase in EToV replication or a deficient assembly.
- Viruses typically proliferate in the lumen of Golgi cisternae. The nucleocapsid tubules approach and attach laterally to the Golgi membrane with one of their rounded extremities facing the membrane.
- The nucleocapsid appears to be stabilized during budding, resulting in a higher electron density and a constant diameter (23 nm).
- Release into the intestinal lumen likely occurs via pinocytosis in reverse. Virus maturation appears to occur intracellularly during the egress process, when the morphology of the virus nucleocapsid transforms from a straight rod (intracellular) to a torus (extracellular).
- The characteristic torus morphology of BToV is observed only in extracellular viral particles or vacuoles close to the cell surface, but never in the cytoplasm.
- Attachment of the viral S protein (and possibly HE, if present) to host receptors mediates virus endocytosis into the host cell.
- Synthesis and proteolytic cleavage of the polyprotein replicase.
- ssRNA(+) genome is released into the cytoplasm upon fusion of the virus membrane with the endosomal membrane (likely mediated by E2).
- Synthesis and proteolytic cleavage of the polyprotein replicase.
- Replication takes place within viral factories. Synthesis of a dsRNA genome from genomic ssRNA(+).
- The dsRNA genome is replicated and transcribed, producing viral mRNAs and new ssRNA(+) genomes.
- Synthesis of subgenomic mRNA-encoded structural proteins.
- Assembly and bud formation at membranes of the endoplasmic reticulum (ER), intermediate compartments, and/or Golgi complex.
- The release of new viruses through exocytosis.
Pathogenesis of Torovirus
- All strains of BToV are pathogenic, causing diarrhea ranging from mild to copious in experimentally and naturally infected young calves.
- Twenty-four to seventy-two hours after exposure, the first clinical symptoms (mild fever, depression, weakness, and anorexia) are observed, followed by 3–5 days of greenish-yellow to brilliant yellow watery diarrhea.
- Calves are susceptible to severe dehydration and death. 24–72 hours after infection, concurrent with the onset of diarrhea, fecal virus is excreted for 2–6 days. Peak shedding occurs three to four days after infection.
- Mixed infections with other enteric viruses, such as rotaviruses or astroviruses, cause diarrhea that is more severe than that caused by either virus alone. In general, CD calves with normal intestinal flora experience more severe diarrhea than Gn heifers.
- BToV can discharge intermittently and frequently for up to four months. Additionally, BToV has been detected in nasal samples. BToV replication and dissemination in the respiratory tract, as well as its role in respiratory pathologies, require additional investigation.
- Horses seroconvert to EToV between the ages of 10 and 12 months without exhibiting symptoms. There have been reports of experimentally infected (intravenous route) animals seroconverting without clinical symptoms. No experiments involving oral infection in horses have been reported to date.
- Multiple studies have demonstrated a correlation between HToV infection and diarrhea in infants. In one study, 35% of infants with enteritis shed HToVs in their feces, compared to 14% of asymptomatic controls (statistically significant difference – odds ratio of 3.1).
- Neither fever nor the presence of other enteric pathogens was noted in the affected children. Recently, HToV excretion in neonates with necrotizing enterocolitis (NEC) has been linked to nosocomial infections. Following HToV infection, immunocompromised children appear to be particularly susceptible to disease.
Epidemiology of Torovirus
- Multiple epidemiological studies have revealed a high seroprevalence of BToV antibodies in a variety of cattle populations, indicating that the virus may circulate frequently in these populations. ELISA and/or RT-PCR have also detected BToV in cattle cases of gastroenteritis.
- Up to 44% of BToV-positive isolates from these cases lacked other significant enteric pathogens. Calves up to 4 months of age are highly susceptible to BToV-induced diarrhea, particularly those younger than 3 weeks.
- The virus has also been identified in five- to six-month-old cattle calves entering sales barns. Calves can experience intermittent BToV discharge during their first 10 months of life.
- BToV can also be excreted at various ages by older calves and adult animals, possibly due to intermittent subclinical infections or the acquisition of new BToV infections.
- The clinical outcome of the infection is influenced by the levels of maternal BToV-specific antibodies circulating in the calf; a seronegative neonatal calf is approximately seven times more likely to develop diarrhea than a seropositive calf.
Transmission and Tissue Tropism of Torovirus
- It has been hypothesized that BToV is transmitted via the oral/nasal route through direct contact with defecation or nasopharyngeal secretions. Under experimental conditions, oral inoculation of calves with BToV has been shown to induce diarrhea with virus secreted in feces.
- BToV antigen and viral RNA have been detected in the nasal secretions of feedlot calves, indicating that the nasal route is another possible entry point. Additionally, intranasal inoculation of Gn and CD heifers has produced diarrhea.
- It has been reported that bovine coronavirus (BCoV) respiratory tract infections occur anterior to enteric infections, indicating the potential importance of this route of transmission in the spread and pathogenesis of this distantly related group of enteric nidoviruses.
- It is feasible that BToV, like BCoV, could replicate and multiply in nasal epithelial cells before being swallowed and infecting the intestines. This hypothesis for the pathogenesis of BToV should be investigated further.
- BToV targets enterocytes from the lower half of the villi extending into the crypts, affecting the caudal portion of the small intestine (mid-jejunum to ileum) and the large intestine. BToV infection of other cell types and organs has not been reported.
Diagnosis of Torovirus
The diagnosis of Torovirus infection involves laboratory testing of samples from the infected individual. Stool samples or respiratory secretions may be collected for analysis.
- Stool sample testing: A stool sample is collected and sent to a laboratory for analysis. The sample is tested for the presence of Torovirus RNA using molecular techniques such as polymerase chain reaction (PCR).
- Respiratory secretion testing: In cases where Torovirus infection is suspected to be respiratory in nature, respiratory secretions such as nasal swabs or throat swabs may be collected for analysis. The samples are tested for the presence of Torovirus RNA using PCR.
- Serological testing: Serological testing may also be used to diagnose Torovirus infection. This involves testing for the presence of antibodies against the virus in blood samples. However, serological testing may not be useful in acute infections as it may take several weeks for antibodies to develop.
Treatment of Torovirus
- There is no specific treatment for Torovirus infection as it is a viral illness. Treatment primarily involves supportive care to manage the symptoms and complications that may arise.
- For mild cases of Torovirus infection, treatment may simply involve rest, hydration, and fever control. Over-the-counter medications like acetaminophen or ibuprofen may be recommended to alleviate fever and pain.
- In more severe cases, hospitalization may be necessary to provide intravenous fluids and electrolytes to prevent dehydration. In some cases, anti-nausea or anti-diarrheal medications may also be prescribed to manage gastrointestinal symptoms.
- It is important to note that antibiotics are not effective against Torovirus infection as it is a viral illness. Additionally, anti-diarrheal medications should be used with caution as they can prolong the duration of the infection by slowing the elimination of the virus from the body.
- Prevention is key to avoiding Torovirus infection. Good hygiene practices such as washing hands thoroughly with soap and water, especially after using the toilet or before eating, can help to reduce the risk of infection. Additionally, avoiding contaminated food and water sources can also reduce the risk of Torovirus infection.
Prevention and Control
There are no specific preventative measures for this virus; however, general hygiene, biosecurity measures, and the consumption of sufficient quantities of colostrum can be used to prevent BToV infections. There are no reports regarding the effects of sterilization or disinfection on toroviruses.
Prevention and control of Torovirus involves adopting good hygiene practices and implementing measures to reduce the risk of transmission.
- Good hygiene practices: Washing hands thoroughly with soap and water, especially after using the toilet or before eating, is essential to prevent the spread of Torovirus. Hands should be washed for at least 20 seconds, using warm water and soap, and dried with a clean towel.
- Food safety: Torovirus can be transmitted through contaminated food or water. It is important to practice food safety by cooking food thoroughly, washing fruits and vegetables before consumption, and avoiding consuming food that has been left out at room temperature for an extended period of time.
- Environmental hygiene: Regular cleaning and disinfection of surfaces and objects that may be contaminated with Torovirus can help to prevent the spread of the virus. This is particularly important in healthcare settings and other high-risk environments.
- Isolation and quarantine: Individuals who have been infected with Torovirus should be isolated to prevent transmission to others. Those who have been exposed to the virus may be placed under quarantine to prevent the spread of the infection.
- Immunization: Currently, there is no vaccine available to prevent Torovirus infection. However, research is ongoing to develop a vaccine.
- Awareness and education: Public awareness campaigns and education about Torovirus can help to raise awareness about the risk of infection and promote good hygiene practices.
FAQ
What is Torovirus?
Torovirus is a genus of single-stranded RNA viruses that can infect both animals and humans.
How is Torovirus transmitted?
Torovirus can be transmitted through fecal-oral contact or respiratory secretions. It can also be spread through contaminated water or food.
What are the symptoms of Torovirus infection in humans?
Symptoms of Torovirus infection in humans can include diarrhea, vomiting, abdominal pain, fever, and respiratory symptoms such as cough and sore throat.
Can Torovirus be treated with antibiotics?
No, Torovirus cannot be treated with antibiotics as it is a viral infection. Treatment usually involves supportive care such as fluid replacement and fever control.
How long does it take for Torovirus symptoms to appear after infection?
The incubation period for Torovirus can range from 12 to 48 hours.
Is there a vaccine for Torovirus?
There is currently no vaccine for Torovirus.
How is Torovirus diagnosed?
Torovirus can be diagnosed through laboratory testing of stool samples or respiratory secretions.
Can Torovirus be prevented?
Torovirus can be prevented through good hygiene practices such as washing hands thoroughly with soap and water, especially after using the toilet or before eating.
Can animals be infected with Torovirus?
Yes, Torovirus can infect a variety of animals including cows, pigs, and horses.
Is Torovirus a common cause of diarrhea in humans?
Torovirus is not as common as other viruses such as norovirus or rotavirus, but it has been identified as a cause of acute gastroenteritis in humans.
References
- Hoet AE, Horzinek MC. Torovirus. Encyclopedia of Virology. 2008:151–7. doi: 10.1016/B978-012374410-4.00516-1. Epub 2008 Jul 30. PMCID: PMC7148633.
- Horzinek, M. C. (1999). TOROVIRUSES (CORONAVIRIDAE). Encyclopedia of Virology, 1798–1803. doi:10.1006/rwvi.1999.0285
- Hu, Z.-M., Yang, Y.-L., Xu, L.-D., Wang, B., Qin, P., & Huang, Y.-W. (2019). Porcine Torovirus (PToV)—A Brief Review of Etiology, Diagnostic Assays and Current Epidemiology. Frontiers in Veterinary Science, 6. doi:10.3389/fvets.2019.00120
- The Journal of Infectious Diseases, Volume 178, Issue 5, November 1998, Pages 1263–1269, https://doi.org/10.1086/314434
- Ujike M, Taguchi F. Recent Progress in Torovirus Molecular Biology. Viruses. 2021; 13(3):435. https://doi.org/10.3390/v13030435
- Jamieson, F. B., Wang, E. E. L., Bain, C., Good, J., Duckmanton, L., & Petric, M. (1998). Human Torovirus: A New Nosocomial Gastrointestinal Pathogen. The Journal of Infectious Diseases, 178(5), 1263–1269. http://www.jstor.org/stable/30117350
- https://decs.bvsalud.org/en/ths/resource/?id=31691&filter=ths_exact_term&q=torovirus
- https://viralzone.expasy.org/127