- In 1968, the rolling circle model of DNA replication was first proposed. This model describes the DNA replication mechanism in circular plasmids and single-stranded circular DNA of viruses.
- Numerous plasmids replicate independently of chromosomal replication. Numerous circularly closed plasmids reproduce autonomously using a process known as rolling circle replication. Unidirectional replication is another name for rolling circle replication.
- Certain circular DNAs reproduce not by the theta (θ) mode we have just studied, but by the rolling circle replication process.
- The intermediates gave this method the name “rolling circle” because the double-stranded portion of the replicating DNA can be compared to a roll of toilet paper unwinding as it moves across the floor.
- This mechanism is sometimes referred to as the θ mode, to differentiate it from the mode, because this intermediate resembles an upside-down Greek letter σ (sigma).
- The rolling circle mechanism is not limited to single-stranded DNA synthesis. This technique is utilised by certain phages (e.g., λ) to replicate double-stranded DNA.
- During the initial step of λ DNA replication, the phage replicates using the θ mode to produce multiple copies of circular DNA.
- These circular DNAs are not packed into phage particles; instead, they act as templates for the rolling circle synthesis of packaged linear λ DNA molecules.
- Here, the replicating fork resembles that of E. coli DNA replication considerably more, with continuous synthesis on the leading strand (the one moving around the circle) and discontinuous synthesis on the trailing strand.
- Before it is packed, the progeny DNA in λ reaches lengths that are many genomes long.
- Multiple-length DNAs are referred to as concatemers. In order to provide each phage head with one genome’s worth of linear DNA, the concatemer is cut enzymatically at the cos sites bordering each full λ genome on the concatemer.
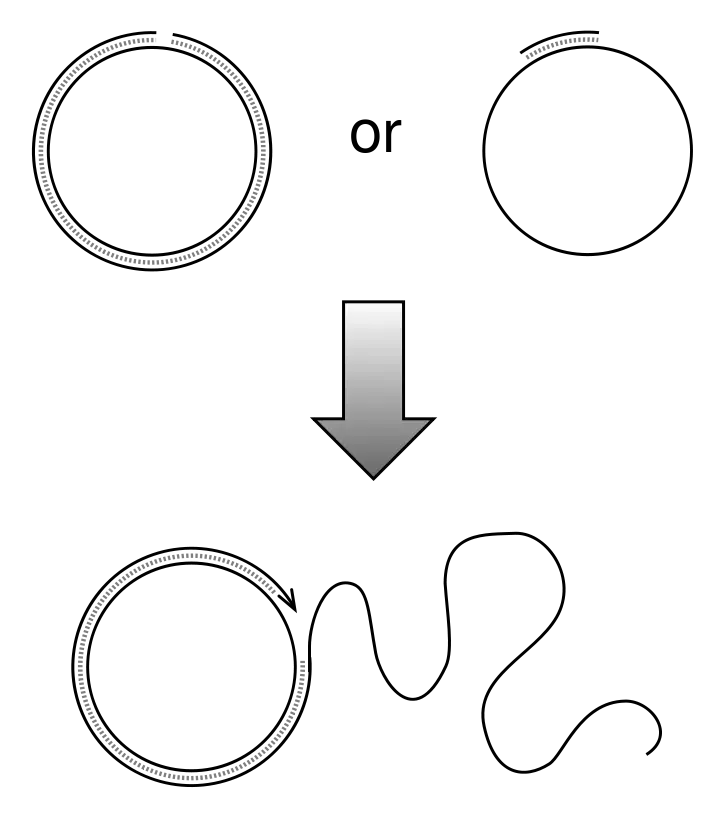
What is Rolling circle replication?
- Rolling circle replication (RCR) is a unidirectional nucleic acid replication mechanism that may rapidly create multiple copies of circular molecules of DNA or RNA, such as plasmids, bacteriophage genomes, and the circular RNA genome of viroids.
- Some eukaryotic viruses also employ the rolling circle process to reproduce their DNA or RNA.
- Rolling circle amplification was designed as a simplified version of natural rolling circle replication, an isothermal DNA amplification process.
- The RCA mechanism is frequently employed in molecular biology and biomedical nanotechnology, particularly in the biosensing field (as a method of signal amplification).
What Is Circular DNA?
- Already known, all living entities have DNA. In general, organisms are divided into two types based on the structure of the cell’s nucleus:
- Eukaryotic Organisms: Their cells contain a well-developed nucleus and various membrane-bound organelles, such as mitochondria, chloroplasts, lysosomes, and endoplasmic reticulum, among others. 80S ribosome are found in eukaryotic cells. Example: human, plants, animals, fungi etc.
- Prokaryotic Organisms: Instead of a formed nucleus, prokaryotic organisms have circular DNA. They lack organelles with membrane-bound components. They contain ribosome 70S. Example: Bacteria, Archaea etc.
- Prokaryotic creatures, like eukaryotes, have double-stranded DNA as their genetic material. In contrast to eukaryotes, bacterial DNA is circular. The DNA of this kind forms a closed loop.
- This form of DNA is also present in mitochondria and plastids of eukaryotic cells, allowing them to divide autonomously.
- DNA replication is a biological process that, according to molecular biology, produces two identical DNA molecules from a single original DNA molecule.
- DNA replication occurs in all living creatures, facilitating cell division. It is the foundation of genetic inheritance.
- The Rolling Circle Mechanism copies circular DNA. DNA replication in eukaryotes is bidirectional. However, the Rolling Circle Mechanism has just one direction.
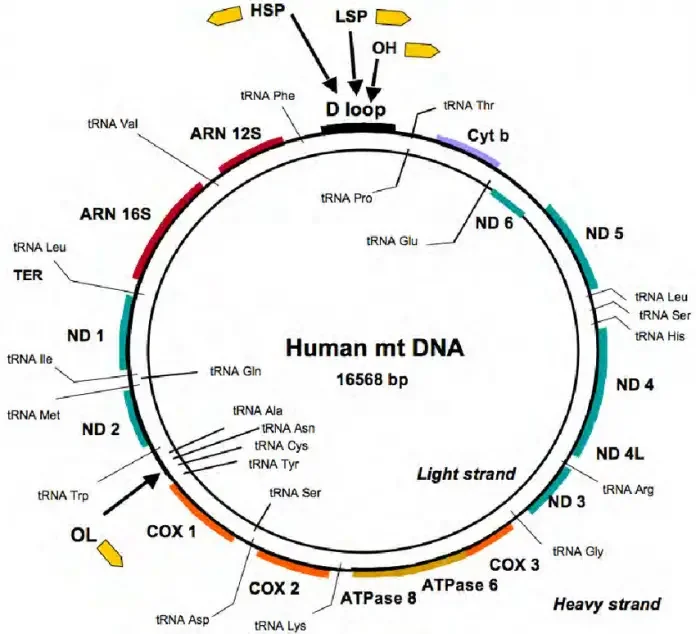
Mechanism of Rolling circle replication
The Rolling circle replication is completed in the following steps;
- Initiation
- Elongation
- Termination
1. Initiation of Rolling circle replication
- The Going Round A protein called nicking enzyme starts the process of DNA replication (RepA).
- This protein is written in plasmid or bacteriophage DNA, which cuts one strand of DNA at a place called “Double-Strand Origin” (DSO).
- Don’t forget that the other strand stays the same (no nicking).
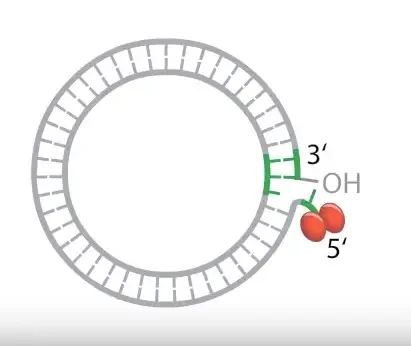
2. Elongation of Rolling circle replication
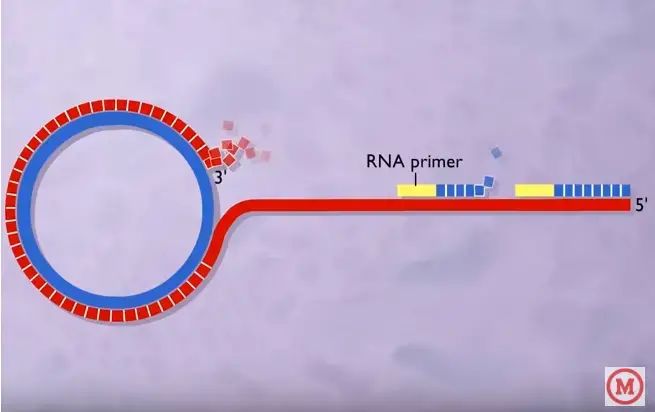
- As shown in the figure, the initiator protein stays attached to the 5’phosphate end of the nicked strand, and the 3’hydroxyl end is freed to act as a primer for the DNA polymerase enzyme.
- This means that the 3′ end is a primer.
- We know that a primer is a short piece of RNA that is used by DNA polymerase to start the process of making copies of DNA. It is needed because the DNA polymerase enzyme can’t put the complementary nucleotides in the 5′ end. It can copy itself in the 3′ end.
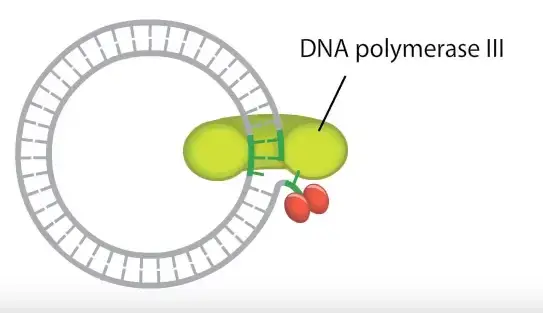
- So, a primer always has both a 5′ end and a 3′ end. DNA polymerase starts the replication process by adding each pair of complementary nucleotides to the 3′ end.
- Now, since the 3′ end is already there, there is no need to add a primer. The nick on the 3′ end makes it work as a primer.
- At the same time, right after the nick is made, an enzyme called DNA polymerase attaches itself to the complementary strand (which is not nicked or the inner circular strand).
- In the presence of Plasmid Replication Initiation Protein, a helicase called PcrA (Plasmid Copy Reduced) that is made by the host moves the nicked strand.
- As DNA synthesis goes on, it can make multiple single-stranded copies of the original DNA in a series called a “Concatemer” that goes from head to tail.
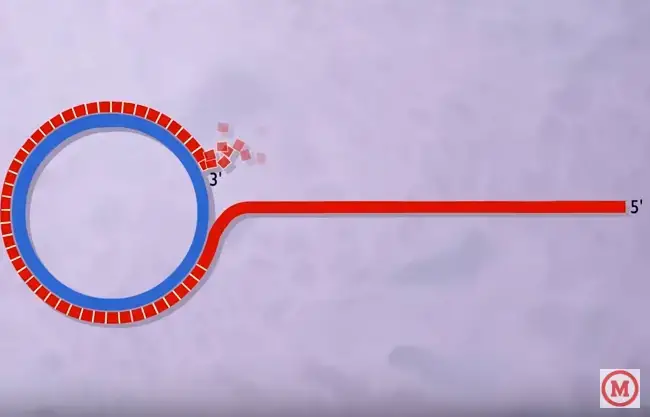
3. Termination of Rolling circle replication
- In this step, copies of the original DNA molecule that are made in a straight line are changed into copies that are made in a circle.
- First, the initiator protein makes another cut in the first (Leading) strand to stop the process of making it (the blue one). So, the first circle is now finished.
- In order to make DNA from the single strand (the red one), RNA polymerase and DNA polymerase III copy the SSO DNA to make a new double-stranded circle.
- Then DNA polymerase I takes out the primer and puts DNA there instead.
- DNA ligase joins the ends together to make a new double-stranded circular DNA molecule.
- The Rolling Circle Mechanism has been used successfully to make more DNA from very small amounts of starting material.
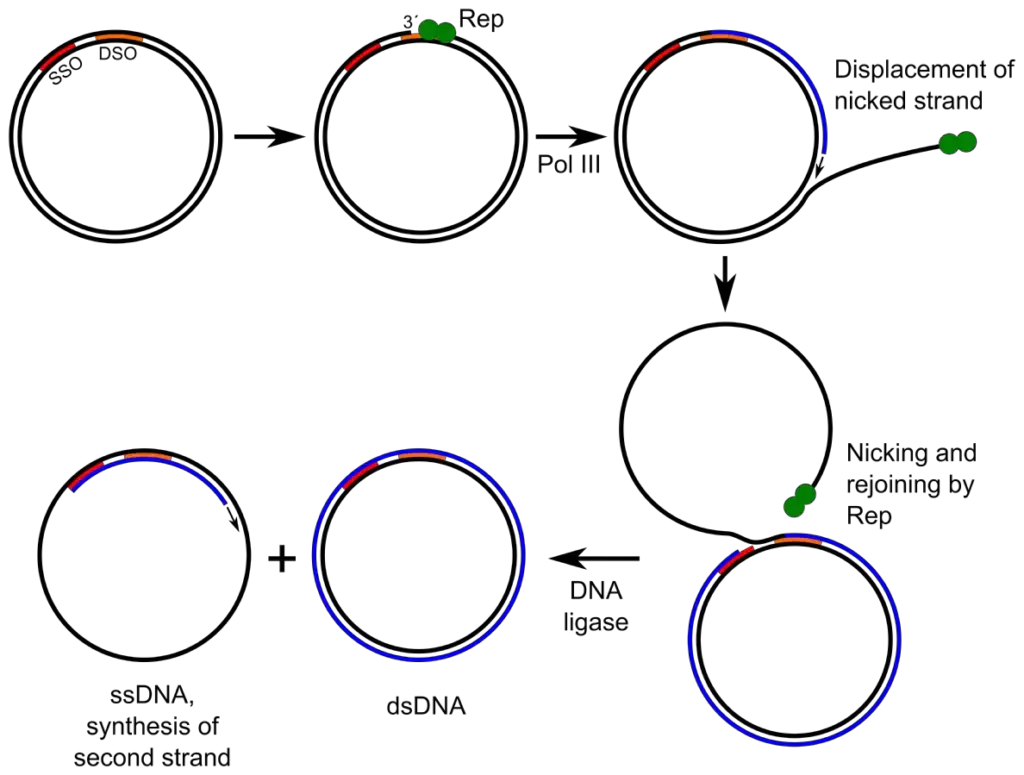
As a summary, a typical DNA rolling circle replication has five steps
- The dsDNA in circular form will be “nicked.”
- Using “unnicked” DNA as the leading strand (template), the 3′ end is lengthened; the 5′ end is displaced.
- A succession of Okazaki fragments are used to convert a lagging strand of DNA into a double-stranded molecule.
- Both “unnicked” and misplaced ssDNA replication.
- Displaced DNA circularises.
Replication of viral DNA using Rolling circle replication
- Some DNA viruses replicate their genomic information in the cells of their hosts by rolling circle replication. Human herpesvirus-6 (HHV-6)(hibv), for instance, encodes a group of “early genes” that are considered to be involved in this process.
- The resulting lengthy concatemers are then cleaved between the pac-1 and pac-2 sections of the HHV-6 genome by ribozymes during packaging of individual virions.
- Human Papillomavirus-16 (HPV-16) is another virus that uses rolling replication to rapidly create offspring.
- Human epithelial cells are infected by HPV-16, which contains a double-stranded circular genome. During replication, the E1 hexamer wraps around the single strand DNA and moves in the 3′ to 5′ direction at the origin.
- In normal bidirectional replication, the two replication proteins dissociate at the time of collision; however, it is suspected that in HPV-16, the E1 hexamer does not dissociate, resulting in continuous rolling replication.
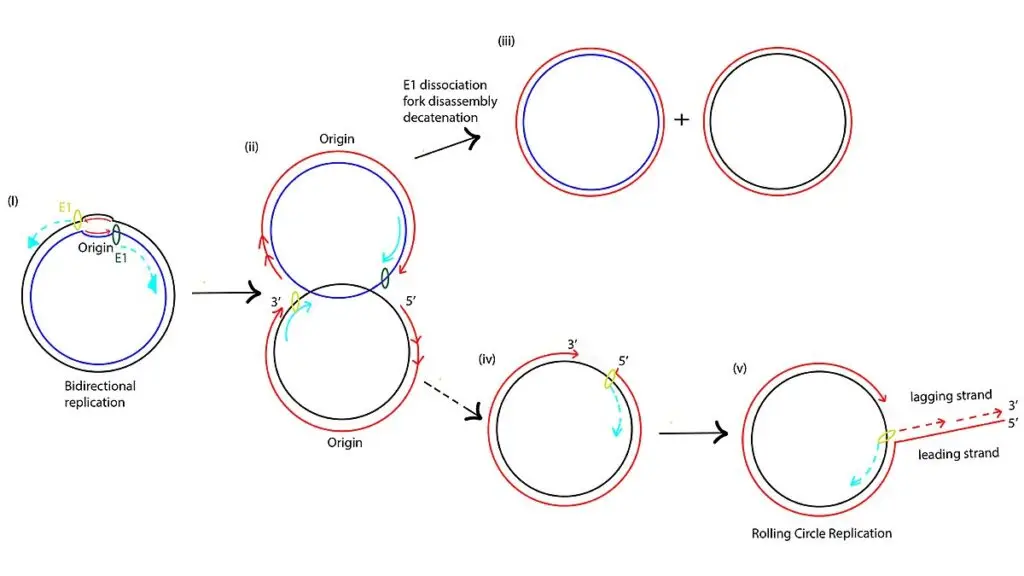
- It is considered that this HPV replication process may have physiological effects on the integration of the virus into the host chromosome and the development of cervical cancer.
- In addition, geminivirus employs rolling circle replication as its mechanism of reproduction. It is a virus that destroys several important crops, including cassava, cotton, legumes, maize, tomato, and okra.
- The DNA of the virus is circular and single-stranded; it replicates in plant cells. Rep, the geminiviral replication initiator protein, is also responsible for modifying the host environment in order to function as part of the replication machinery.
- The presence of motifs I, II, and III at its N terminus makes Rep strikingly comparable to the majority of rolling replication initiation proteins of eubacteria.
- During rolling circle replication, geminivirus ssDNA is transformed to dsDNA, and Rep is then coupled to the dsDNA at the origin sequence TAATATTAC.
- After Rep and other replication proteins bind to dsDNA, a stem loop is formed and the DNA is cleaved at the nanomer sequence, producing strand displacement.
- This displacement permits the replication fork to advance in the 3′ to 5′ direction, resulting in the formation of a new ssDNA strand and a concatameric DNA strand.
- The intermediates of bacteriophage T4 DNA replication consist of circular and branched circular concatemeric structures.
- These formations likely reflect a reproduction mechanism involving a rolling circle.
Replication of viral RNA using Rolling circle replication
- Some RNA viruses and viroids reproduce their genomes using rolling circle RNA replication.
- There are two different RNA replication methods for viroids, followed by members of the families Pospivirodae (asymmetric replication) and Avsunviroidae (symmetric replication) (symmetric replication).
- A host RNA polymerase converts the circular plus strand RNA in the family Pospiviroidae (PSTVd-like) into oligomeric minus strands and then oligomeric plus strands.
- These oligomeric plus strands are broken by the host RNase and ligated by the host RNA ligase to produce monomeric plus strand circular RNA. This is known as the asymmetric rolling circle replication process.
- The viroids belonging to the family Avsunviroidea (ASBVd-like) reproduce their genome using the symmetric rolling circle replication route.
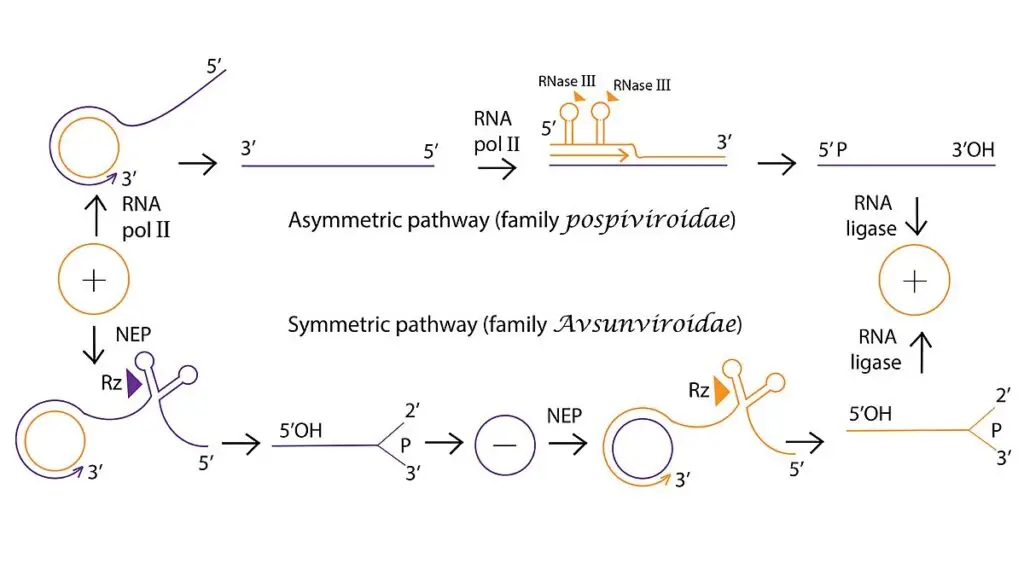
- In this symmetric process, oligomeric minus strands are cleaved and ligated into monomeric minus strands before being transcribed into oligomeric plus strands.
- These oligomeric plus strands are subsequently broken and ligated to produce a monomeric plus strand.
- The symmetric replication route was given its name because both positive and negative strands are created in the same manner.
- The self-cleaving hammerhead ribozyme structure in the Avsunviroidae splits the oligomeric plus and minus strands, but there is no such structure in the Pospiviroidae.
What is Rolling circle amplification?
The derivative form of rolling circle replication has been successfully applied to the amplification of DNA from very modest initial quantities. This method of amplification is known as Rolling circle amplification (RCA).
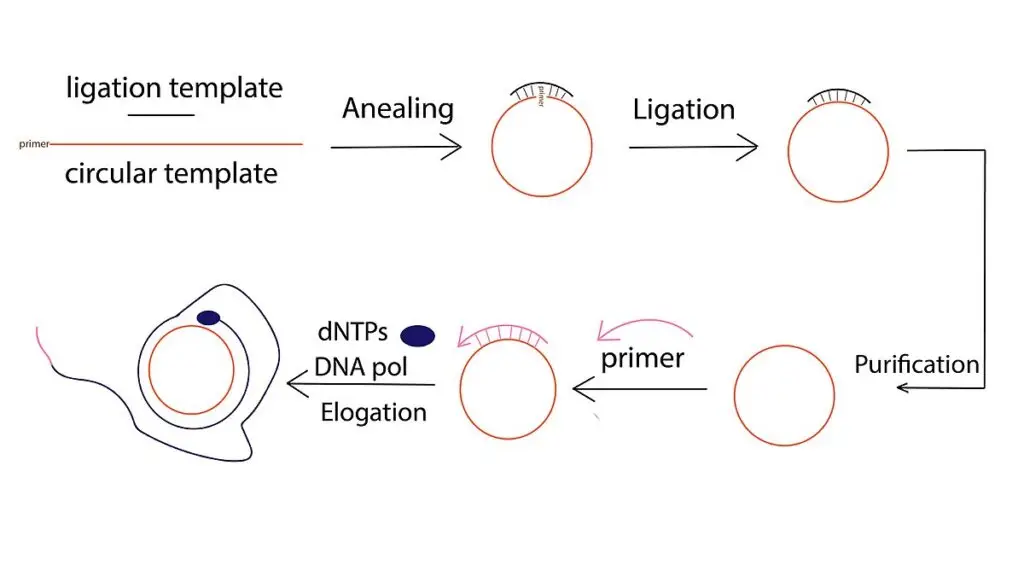
- RCA is an isothermal nucleic acid amplification process in which the polymerase constantly adds single nucleotides to a primer annealed to a circular template, producing a lengthy concatemer ssDNA containing tens to hundreds of tandem repetitions (complementary to the circular template).
Components Required for Rolling circle amplification
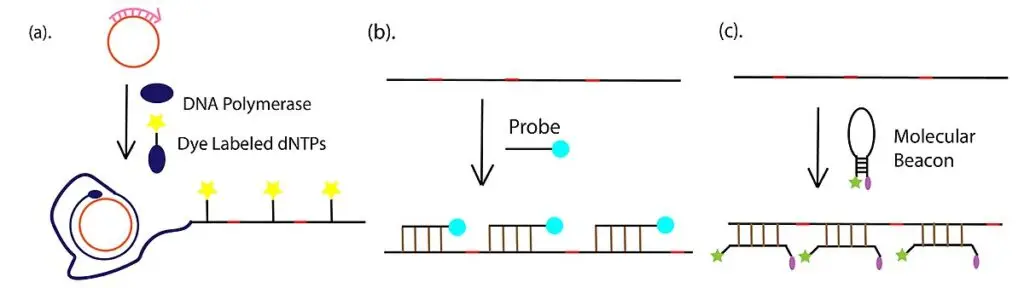
Five essential components are required to perform an RCA reaction:
- A DNA polymerase
- A compatible buffer with the polymerase.
- A brief DNA or RNA starter
- A circular DNA template
- Triphosphates of deoxynucleotide (dNTPs)
Phi29, Bst, and Vent exo-DNA polymerase are employed in RCA for DNA amplification, and T7 RNA polymerase is used for RNA amplification. Since Phi29 DNA polymerase has the highest processivity and strand displacement ability among the aforementioned polymerases, it has been utilised in RCA reactions the most frequently. Unlike polymerase chain reaction (PCR), RCA can be done at a constant temperature (from room temperature to 65 degrees Celsius) in both free solution and on immobilised targets (solid phase amplification).
Steps of DNA RCA reaction
Typically, there are three steps to a DNA RCA reaction:
- Circular template ligation, which can be carried out by template-mediated enzymatic ligation (e.g., T4 DNA ligase) or template-free ligation with specialised DNA ligases (i.e., CircLigase).
- elongation of a single DNA strand caused by a primer. It is possible to use multiple primers to hybridise with the same circle. Thus, several amplification events can be initiated, resulting in multiple RCA products (“Multiprimed RCA”).
- Fluorescent detection is typically used for amplification product detection and visualisation, utilising fluorophore-conjugated dNTP, fluorophore-tethered complements or fluorescently-labeled molecular beacons. In addition to fluorescence techniques, gel electrophoresis is also commonly employed to detect RCA products.
RCA results in the linear amplification of DNA, since each circular template expands at a predetermined rate over a predetermined period of time. Several methods have been investigated in an effort to boost yield and achieve exponential amplification, like PCR does. One of these is hyperbranched rolling circle amplification, or HRCA, in which primers that anneal to the initial RCA products are added and prolonged.
In this manner, the original RCA develops more amplifiable templates. Another technique is circle to circle amplification, or C2CA, in which the RCA products are digested with a restriction enzyme and ligated into new circular templates using a restriction oligo, followed by a second round of RCA with a greater quantity of circular templates for amplification.
Applications of Rolling circle replication (RCA)
- RCA can multiply a single molecular binding event over a thousandfold, making it especially helpful for identifying ultra-low abundance targets.
- In addition to free solution environments, RCA reactions can be conducted on solid surfaces such as glass, micro- or nano-beads, microwell plates, microfluidic devices, and even paper strips. This characteristic makes it a highly effective instrument for signal amplification in solid-phase immunoassays (e.g., ELISA). In this manner, RCA is becoming a very adaptable signal amplification technique with numerous applications in genomics, proteomics, diagnostics, and biosensing.
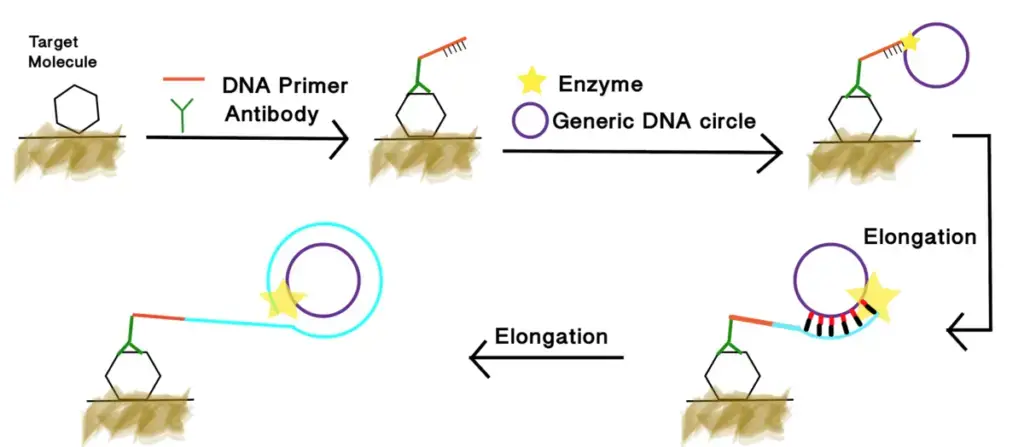
- Immuno-RCA is an isothermal signal amplification technique for the detection and quantification of proteins with high specificity and sensitivity. This technology combines two distinct fields: RCA, which permits nucleotide amplification, and immunoassay, which employs antibodies specific to intracellular or free biomarkers. Immuno-RCA provides a specific signal amplification (high signal-to-noise ratio), making it excellent for detecting, measuring, and visualising low abundance proteic markers in liquid-phase immunoassays and immunohistochemistry.
- In the field of biosensing, numerous RCA derivatives were widely used. RCA has been used successfully to detect viral and bacterial DNA in clinical samples, which is extremely advantageous for the quick diagnosis of infectious disorders.
- It has also been implemented as an on-chip signal amplification technique for nucleic acid (DNA and RNA) microarray assays.
References
- Roy, Suvam & Sengupta, Supratim. (2021). Evolution towards increasing complexity through functional diversification in a protocell model of the RNA world. 10.1101/2021.05.10.443418.
- Filutowicz, M. (2009). Plasmids, Bacterial. Encyclopedia of Microbiology, 644–665. doi:10.1016/b978-012373944-5.00014-6
- https://www.biotechfront.com/2021/06/rolling-circle-model-of-replication.html
- https://plantlet.org/replication-of-circular-dna-rolling-circle-model/
- https://viralzone.expasy.org/2676
- https://www.onlinebiologynotes.com/mechanism-of-plasmid-replication-theta-and-rolling-circle-dna-replication/
- https://www.bionity.com/en/encyclopedia/Rolling_circle_replication.html
- https://www.biologydiscussion.com/dna/dna-replication/rolling-circle-method-of-dna-replication-genetics/67533
- https://www.morebooks.de/shop-ui/shop/product/978-613-8-87580-2
- https://biocyclopedia.com/index/genetics/chemistry_of_the_gene_synthesis_modification_and_repair_of_dna/rolling_circle_model_of_dna_replication.php
- https://en.wikipedia.org/wiki/Rolling_circle_replication
- https://www.slideshare.net/HannanZoologist/rolling-circle-model-of-dna-replication
- https://teaching.ncl.ac.uk/bms/wiki/index.php/Rolling_circle_replication