What is Real-Time PCR?
- Real-Time PCR (also called quantitative PCR or qPCR) is a variation of the Polymerase Chain Reaction which allows the progress of DNA amplification to be monitored during each cycle, rather than only at end point.
- The fluorescence signal is used as reporter, which increases proportionally with the amount of PCR product (amplicon) present in reaction, and is measured in real time.
- The method employing fluorescent dyes (for example SYBR Green) or sequence-specific probes (for example TaqMan) is used, depending on requirement of specificity and sensitivity.
- The starting quantity of nucleic acid (DNA or RNA after reverse transcription) is inferred by cycle number at which fluorescence crosses a defined threshold (Ct or Cq), fewer cycles indicating greater initial template.
- Advantages are provided by Real-Time PCR in terms of sensitivity, speed, dynamic range, and reduced risk of contamination since no post-PCR manipulation (eg gel electrophoresis) is needed.
- When RNA is target, reverse transcription is performed first to convert RNA into complementary DNA (cDNA) then amplification / detection by qPCR is done.
- The instrument used must have capability of fluorescence detection and thermal cycling, so that fluorescence data can be collected after each cycle.
- Applications are many (diagnostics, gene expression quantification, detection of pathogens etc), because Real-Time PCR allows both qualitative (presence/absence) and quantitative (copy number, relative expression) analysis.
Definition of Real-Time PCR
Real-time PCR, also known as quantitative PCR (qPCR), is a molecular biology technique that allows for the amplification, detection, and quantification of DNA or RNA in real-time during the amplification process. It uses fluorescent dyes or probes to measure the increase in fluorescence intensity, providing quantitative data on the amount of target nucleic acid present in the sample.
Principle of Real-Time PCR – How Does Real-Time PCR Work?
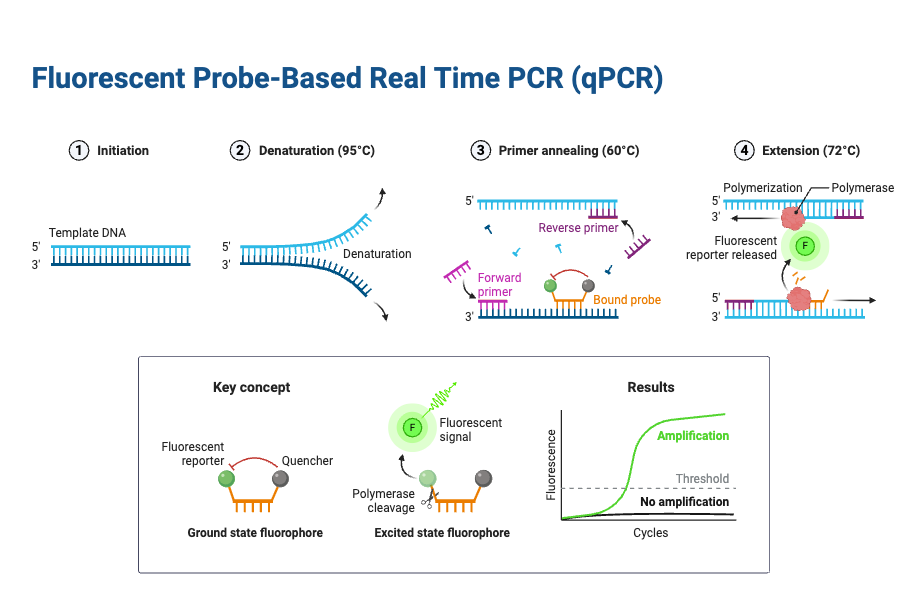
The Principle of Real-Time PCR is based on amplification of target DNA (or cDNA) by Thermal Cycling, while fluorescence is monitored continuously, so that product accumulation is detected during each cycle.
With inclusion of Reporter / fluorescent dye (non-specific dyes like SYBR Green, or sequence-specific probes such as TaqMan), the fluorescence signal increases in proportion to the amount of PCR product produced, and this increase is measured cycle by cycle.
The conversion of RNA into complementary DNA (cDNA) is performed first, when RNA is to be analysed, because DNA polymerase cannot amplify RNA directly; then the PCR amplification proceeds using cDNA as template.
During each cycle, three main temperature-dependent steps are performed: denaturation (strand separation), annealing (primer binding), extension (new strand synthesis by DNA polymerase), as in conventional PCR; but with the added detection stage via fluorescence after or during the extension or probe cleavage step.
A threshold fluorescence level is defined, above background noise, and the cycle number at which fluorescence crosses this threshold (Cq or Ct value) is used to infer the initial quantity of target nucleic acid, because earlier crossing indicates larger starting template.
Amplification efficiency is assumed (or must be validated) to be approximately constant during exponential phase, so that doubling (or near doubling) of product per cycle occurs until reaction components become limiting, then plateau is reached.
The instrument (thermal cycler with optical-detection module) is equipped to both change temperature rapidly for each PCR step, and detect fluorescence (via detector / camera etc) in a closed-tube format, so that signal is collected without opening reaction vessels (which reduces contamination).
Melting curve / dissociation curve analysis can be applied after amplification when non-specific dyes are used, to verify specificity of the product, by observing at which temperature double-stranded DNA melts (dissociates) which reflects product sequence purity.123
Steps of Real-Time PCR (Protocol)
- At first, RNA is extracted from cells / tissues, which must be purified and free of contaminants such as proteins, genomic DNA, RNases.
- After extraction, RNA integrity is checked, often by spectrophotometry (A260/A280) or agarose gel / Bioanalyzer, so that only good quality RNA is used.
- Genomic DNA is removed, by DNase treatment or other methods, so that amplification of contaminating DNA is avoided in downstream steps.
- Reverse Transcription (RT) is performed, in which RNA is converted into complementary DNA (cDNA), using Reverse Transcriptase enzyme, suitable primers (gene-specific / random / oligo-dT), dNTPs, buffer, and co-factors.
- The RT reaction is incubated at appropriate temperature (often ~42-50°C) for required time (for example 30-60 min), so that reverse transcription proceeds; then RT is inactivated by heat if needed.
- Real-Time PCR mix is prepared, by combining cDNA template, forward and reverse primers, DNA polymerase (thermostable), fluorescent reporter (dye or probe), buffer, Mg2+ etc. Master mix may be prepared to reduce pipetting variation.
- Thermal cycling is performed: denaturation, annealing, extension steps repeated for many cycles (often ~35-45), with fluorescence measurement at each cycle.
- Fluorescence detection is done in each cycle (either during extension or after), via a dye (eg SYBR Green) or probe (eg TaqMan), so that accumulation of PCR product is monitored in real time.
- Threshold is set above background fluorescence, and cycle number at which fluorescence crosses threshold (Ct or Cq) is determined, which is used for quantification of initial template amount.
- Controls are included: no-template control (NTC) to check for contamination, no-RT control to check for genomic DNA contamination, positive control / housekeeping gene to check assay performance.
- After amplification, when using non-specific dyes, melting curve (dissociation curve) analysis is performed, so that specificity of product is verified (to detect primer dimers or non-specific amplification)
- Data are analysed, Ct / Cq values are compared across samples, efficiencies may be checked, normalization done (with internal control genes), relative or absolute quantification is performed.
The working procedure of Real-Time PCR
The working procedure of Real-Time PCR is generally divided into two broad steps, and these steps are amplification and detection.
A. Amplification
- Denaturation – At very high temperature (usually around 95 °C), the double-stranded DNA is separated into single strands, and secondary structures are loosened. The maximum temperature which the DNA polymerase can withstand is usually chosen, although when GC content is high, the denaturation time is sometimes increased for better efficiency.
- Annealing – In this step, the primers hybridize with complementary sequences. An appropriate temperature is selected, usually about 5 °C lower than the melting temperature (Tm) of the primers, so that specific binding occurs.
- Extension – At temperature near 70–72 °C, DNA polymerase shows its optimal activity, and primer extension is carried out, often at a rate of almost 100 bases per second. In cases where the amplicon is very small, the extension step is sometimes combined with annealing step at around 60 °C, which reduces cycle time.
B. Detection
- Fluorescence principle – The detection part is carried out with fluorescence technology, which is added in the reaction in the form of dyes or probes.
- Thermal cycling – The specimen is placed into the proper wells and subjected to thermal cycles, like in conventional PCR, but here, the detection is simultaneous.
- Light source – A tungsten or halogen source is applied by the Real-Time PCR machine, which excites the marker dye, and fluorescence is produced in direct proportion with copy number increase.
- Signal emission – The emitted fluorescence is captured by the detector, which then converts the optical signal into digital form. After that, data is sent into the computer system.
- Threshold crossing – The signal is observed on a screen, and when fluorescence crosses a defined threshold level (minimum detection level of the detector), the sample is considered positive, and the cycle number of that crossing is recorded (Ct value).8
Real-time PCR assay types
Gene expression profiling assays – In such assays, the relative abundance of transcripts is assessed, and expression patterns between different samples are determined. RNA quality, reverse transcription efficiency, and real-time PCR efficiency are considered crucial. The strategy for quantification is selected, and a normalizer gene is chosen, because normalization ensures reliability of comparison. Gene expression assays are widely used but they require careful optimization, since minor variations in cDNA synthesis may produce large changes in calculated expression.
Viral titer determination assays – These assays are designed to quantify viral copy number within samples. Quantification is usually performed by comparison with a standard curve, which is generated using known genome equivalents or nucleic acid extracted from a titered virus control. The reliability of such assays is dependent on the accuracy of the material used in curve generation. When the target is an RNA virus, reverse transcription must be included before PCR, and the efficiency of both steps affects results. The Assay design is also influenced by whether the aim is to quantify functional infectious virus, or total viral particles present.
Genomic profiling assays – In these assays, genomic DNA is examined for duplications or deletions. The design depends strongly on whether relative or absolute quantification is intended, because this determines how the standard curve is constructed. A major focus is placed on the efficiency of real-time PCR, and the precision required to discriminate even single-copy differences in the genome. For many experimental systems, this requirement for accuracy makes optimization challenging and sometimes repetitive.
Allelic discrimination assays – These are employed to detect genetic variation, even at the level of a single nucleotide polymorphism (SNP). Unlike the previously described assays, the endpoint fluorescence is measured instead of the continuous monitoring across cycles. The information obtained is used to determine the zygosity status of a genome. Primer and probe design have critical importance, because allele-specific cross-reactivity must be minimized to avoid misleading signals. In such assays, sensitivity and specificity both are highly dependent upon probe chemistry.
What is one-step RT-PCR?
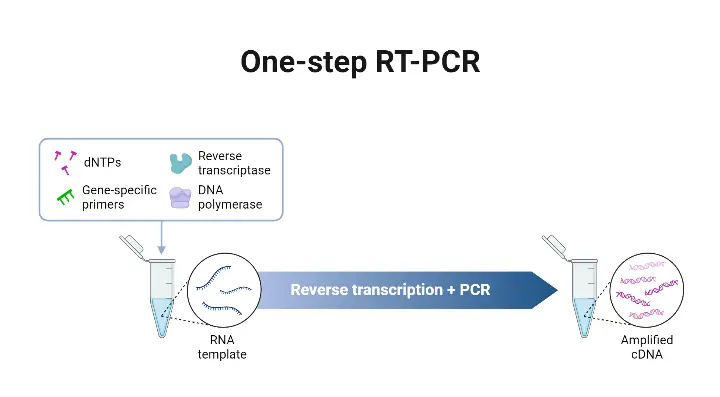
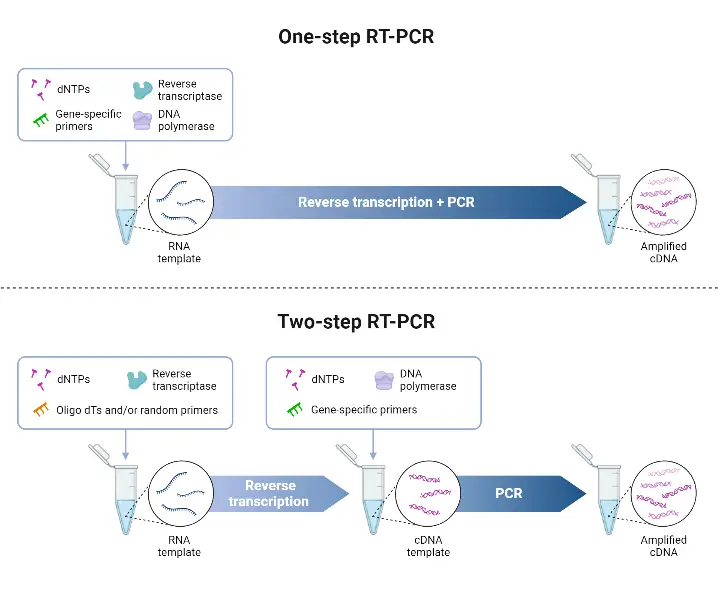
- The One-step RT-PCR method is a protocol in which reverse transcription (RT) and polymerase chain reaction (PCR) are combined in a single reaction tube, in a common buffer, with both reverse transcriptase and DNA polymerase present.
- The RNA is converted into complementary DNA (cDNA) by the reverse transcriptase, and amplification of that cDNA is effected by the DNA polymerase, all without transferring material between tubes.
- The One-step format is chosen when speed, simplicity, or high-throughput are required, because fewer pipetting steps are involved, workflow is simplified, and risk of contamination is reduced.
- Gene-specific primers are required in One-step RT-PCR, since RT and PCR are in same reaction, and non-specific priming (eg oligo(dT) or random hexamers) may not be compatible with PCR conditions that follow.
- It is less flexible than Two-step RT-PCR in terms of target number per sample, because only those targets for which primers are included can be amplified in same tube.
- The sensitivity or efficiency may be somewhat lower in some scenarios, because reaction conditions must compromise between optimal RT and optimal PCR phases.
- The method is advantageous when sample RNA is limited or when fewer genes are to be measured, as material loss is minimized, and turnaround time is shortened.
- In One-step RT-qPCR (quantitative version), the above process is coupled with a detection system (fluorescent probes or dyes) so that the amplified product is measured in “real time” as amplification proceeds.14
What is Two-step qRT-PCR?
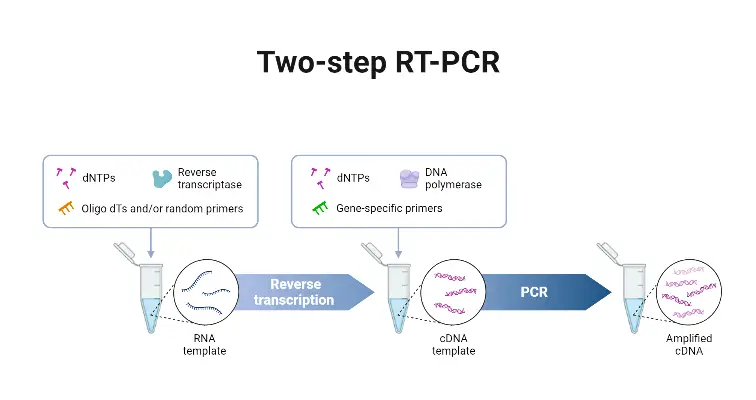
- Two-step qRT-PCR is a technique in which the reverse transcription (RT) and the quantitative PCR (qPCR) are performed in separate reactions.
- The RNA first is transcribed into complementary DNA (cDNA) by reverse transcriptase, using primers such as oligo(dT), random hexamers, or gene-specific primers.
- Then, an aliquot of that cDNA is used as template in a separate real-time PCR reaction to amplify specific target sequences and quantitate them.
- The flexibility in primer choice for the RT step is provided, which allows for broad transcript coverage (when random/oligo(dT) primers are used), or specific targeting (when gene-specific primers are employed).
- Greater sensitivity is often achieved, especially for low abundant transcripts, because each reaction (RT and qPCR) can be optimized independently.
- A stable cDNA pool is generated, which can be stored for later use, or used in multiple qPCR reactions for different targets from the same original RNA sample.
- Disadvantages include increased handling steps, which may raise risk of contamination, more time being required, and more complexity in workflow.
- Use-cases are provided mostly where multiple genes are to be quantified from one RNA sample, or when assay sensitivity / flexibility is needed (eg, challenging sequences or unknown targets).13
Differences Between one-step vs two-step RT-PCR
| Feature | One-step RT-PCR | Two-step RT-PCR |
|---|---|---|
| Definition / Setup | Combined RT + PCR in a single tube, single buffer, with reverse transcriptase + DNA polymerase together. | RT done first (to convert RNA → cDNA) then PCR amplification in a separate reaction using aliquot of cDNA. |
| Primers / Priming Options | Gene-specific primers needed, because all reactions occur together. | Flexible priming: oligo(dT), random hexamers, or gene-specific primers may be used in RT; then PCR with appropriate primers. |
| Flexibility / Multiplexing | Lower flexibility: limited ability to amplify many targets or to re-use cDNA for new/unplanned targets. | Higher flexibility: cDNA produced can be stored, aliquoted, used for many PCRs; targets may be added later. |
| Sensitivity / Efficiency | Sometimes lower sensitivity or compromised efficiency, because RT and PCR conditions must be compromise. | Often higher sensitivity or efficiency, when conditions for RT and for PCR can be optimized independently. |
| Risk of Contamination / Error | Lower risk, because fewer handling steps, fewer tube transfers, less pipetting. | Higher risk, because more handling, more tube transfers, more opportunities for error or contamination. |
| Time / Workflow Complexity | Faster, simpler workflow. Less hands-on time. | More time consuming, more steps to setup, more optimization needed. |
| Reagent Use / Cost | Less reagent usage (smaller reaction sizes, combined enzymes) often lower cost for single target assays. | More reagent usage, more consumables, possibly higher cost especially when multiple targets or experiments are done. |
| Best Use-Case / Application | High-throughput screening when few known targets, when speed and simplicity are important. | Experiments where multiple genes targets are needed, or when sample is limited, or when later targets may need to be added, or when optimization is critical. |
Fluorescence Markers used in Real Time PCR
- The Types of fluorescence markers are broadly divided into DNA-binding dyes (non-specific) and sequence-specific probes, which differ in specificity, cost, design complexity.
- The DNA-binding dye (such as SYBR Green I) is used, which binds to any double-stranded DNA (dsDNA) in the reaction; when bound fluorescence increases strongly, and signal reflects total dsDNA present (target + non-specific products).
- The TaqMan probe chemistry is used, which employs an oligonucleotide probe labelled with a fluorescent reporter at 5’ end and quencher at 3’ end; probe hybridizes to target, and during extension the probe is cleaved by the polymerase’s 5′→3′ nuclease activity, releasing reporter from quencher, signal being produced only if correct target is present.
- Molecular Beacons are used, which are hairpin-shaped probes, with a fluorophore and quencher at ends; fluorescence is quenched when probe is in stem-loop form but restored when probe hybridizes to target sequence, because stem loop opens.
- The Scorpion probes / LUX / Eclipse / hybridization (LightCycler-type) probes are also used, which employ different designs of probe / primer combinations to get specificity, sometimes using fluorescence resonance energy transfer (FRET) or quencher/reporter separation upon binding or extension.
- The Reporter dyes commonly used include FAM, VIC, TET, etc, which differ in excitation/emission spectra, and are used in dual-labelled probes like TaqMan probes.
- The Quencher moieties are included in probe-based systems, which suppress reporter fluorescence until the probe is cleaved or binds target. Examples include TAMRA, DABCYL, or dark quenchers like Black Hole Quencher (BHQ) that do not emit light, minimising background.
- Passive reference dyes are used in many master mixes, which help normalize well-to-well / cycle-to-cycle variability of fluorescence that is not due to amplification (optical differences, pipetting etc).
- dsDNA-binding dyes are cost-effective / simpler design, because only primers are needed, but specificity is lower, and melting-curve analysis is often required to verify correct amplicon.
- With probe-based markers, high specificity is achieved, because only target-specific probe binding or cleavage yields fluorescent signal, reducing false positives coming from primer-dimers or non-target amplifications.5467
Applications of Real-Time PCR
- The Gene expression (mRNA) analysis is performed, because changes in transcript levels under different conditions (drug treatment, stress, developmental stages etc) are quantitated.
- Genetic variation analysis / mutation detection is done, which allows identification of single nucleotide polymorphisms (SNPs), insertions / deletions, or point mutations in genes.
- Pathogen detection and quantification is enabled, because Real-Time PCR can detect (and measure) viral, bacterial, fungal nucleic acid in samples with high sensitivity.
- Diagnostic testing is facilitated, as screening / monitoring of infectious diseases, and genetic disease markers can be done using real-time PCR, often for early detection / disease progression / therapeutic monitoring.
- GMO (Genetically Modified Organism) detection is carried out, because specific transgene or modification sequences are identifiable and measurable in crop / food / environmental samples
- Allelic discrimination and genotyping is applied, so that different alleles in a population or sample are distinguished, which is important in studies of variation or pharmacogenomics.
- MicroRNA and noncoding RNA analysis is supported, because even small non-coding RNA transcripts can be reverse transcribed and quantified, providing insight into regulatory RNAs.
- Copy number variation (gene dosage) is measured, which is beneficial in research where gene amplification / deletion is significant (eg oncology) since gene copy changes impact phenotype.
- Environmental / ecological studies are aided, since microbial load or species abundance can be quantified in environment or food / soil / water samples.
- Monitoring of treatment or therapeutic efficacy is conducted, because real-time PCR allows viral load / pathogen load to be tracked over time, for assessing response to therapy.
- Mutation / variant tracking is executed, especially in epidemiology, so that emerging variants in pathogens (virus variants etc) or mutational shifts are identified.
- Quality control in biomanufacturing or diagnostics is insured, because real-time PCR is used to check purity, contamination, or presence of unwanted nucleic acids.91011
Advantages of Real-Time PCR
- The Dynamic range of detection is broad, which enables quantification across many orders of magnitude (eg 6-8 logs), so both low and high abundance targets are measured precisely.
- High sensitivity is provided, because small amounts of template (DNA or RNA) are detectable, and low copy number targets can be found.
- Quantification is done during exponential phase, which yields more accurate / reliable data than endpoint (plateau) detection, because exponential amplification is less influenced by reagent depletion and non-linear effects.
- The initial copy number of target is allowed to be determined with accuracy, because Ct / Cq values are captured early in reaction.
- Reduced post-PCR handling is achieved, which lowers risk of contamination, because amplification and detection are performed in same closed tube system.
- Rapid turnaround of results is enabled, since gel electrophoresis or other end-point analyses are not required, which saves bench time / hands-on time.
- Multiplexing capability is supported, because multiple fluorescent dyes / probes can be used in same reaction, which allows simultaneous detection of several targets.
- Improved reproducibility is implied, because threshold cycles (Ct/Cq) and standard curves or internal controls are used, which help normalize variation across different runs / samples.
- Throughput is increased, as many samples / wells are processed automatically by real-time PCR machines, and data analysis is often software-assisted.
- Flexibility in qualitative / quantitative outcomes is allowed, because real-time PCR can be used to detect presence/absence of sequences, or to measure how many copies are present.
- Specificity is improved when sequence-specific probes are used, because non-specific signals (primer-dimers, off-target amplifications) are reduced, which improves signal clarity.12
Real-Time vs. Digital PCR vs. Traditional PCR
| Feature | Traditional PCR | Real-Time PCR (qPCR) | Digital PCR (dPCR) |
|---|---|---|---|
| Quantification type | Semi-quantitative / qualitative is produced, because measurement is done at end-point, often by gel electrophoresis. | Quantitative measurement is produced, because fluorescence is monitored during each cycle, and Ct / Cq is used. | Absolute quantification is produced, because sample is partitioned and positive / negative partitions counted, no need of standard curve. |
| Sensitivity & low copy detection | Lower sensitivity is suffered, because endpoint detection is less precise, non-linear plateau phase introduces high variability. | High sensitivity is achieved, because detection during exponential phase allows detection of small fold changes. | Very high sensitivity is enabled, especially for rare targets or low abundance molecules, because partitioning reduces background and enhances detection of rare events. |
| Dependence on standards / calibration | Standard curve or external controls are often required or results are compared qualitatively. | Standard curve or reference samples are required to convert Ct / Cq into absolute or relative quantity. | No standard curve is needed, because absolute quantification is done via counting positives vs negatives and using Poisson statistics. |
| Reaction monitoring | No reaction monitoring during cycles; only final product is seen. | Monitoring during cycles is done (fluorescence each cycle), so exponential phase data is captured | End-point detection in partitions is done (post amplification in partitions), but counting positive partitions gives measure of original quantity. |
| Tolerance to PCR inhibitors / complex samples | Lower, because inhibitors can affect the amplification kinetics thus endpoint detection suffers more. | Moderate; some tolerance depending on reagents, but efficiency issues can impact Ct values. | Higher tolerance, because partitioning isolates some reactions, thus some partitions may be unaffected, improving reliability. |
| Throughput & cost per sample | Lower cost equipment, cheaper reagents; but many downstream handling steps (gel etc) increase labor/time costs. | Moderate to high throughput possible; cost per reaction is higher than traditional, instrument cost is higher, but labor and time savings exist. | Cost per sample is higher, partitioning adds complexity; throughput depends on instrument; higher precision sometimes trades off with cost/time. |
| Use in rare mutation / variant detection | Poor – changes that are subtle or rare are often not detectable reliably. | Good, but detection of very low frequency variants is harder, because standard-curve & sensitivity limitations exist. | Excellent – low frequency / rare variant detection is a major advantage, due to partition approach. |
| Dynamic range | Narrower dynamic range is provided, because endpoint plateau collapses distinctions between sample starting amounts. | Wide dynamic range is obtained, since exponential amplification is measured before plateau. | Also wide dynamic range, though partition limits and sample volume may impose practical constraints. |
| Speed / turnaround | Slower overall workflow, because post-PCR analyses (electrophoresis, staining) are required. | Faster, since detection and quantification are done in real-time, no need for gels etc. | Variable – partitioning step adds some time, but once set up, absolute quantification with minimal downstream work is possible. |
FAQ
What is real-time PCR?
Real-time PCR, also known as quantitative PCR (qPCR), is a molecular biology technique used to amplify and quantify specific DNA or RNA sequences in real time during the PCR process.
How does real-time PCR differ from conventional PCR?
In conventional PCR, amplification results are analyzed after the reaction is completed, whereas real-time PCR allows for continuous monitoring of the amplification process and quantification of the target DNA or RNA during the reaction.
What are the key components required for real-time PCR?
Real-time PCR requires a thermal cycler instrument with the ability to measure fluorescence signals, specific primers for the target sequence, a DNA polymerase enzyme, fluorescent probes or dyes, and a sample containing the target DNA or RNA.
What are the advantages of real-time PCR?
Real-time PCR offers several advantages, including rapid amplification and quantification of target sequences, high sensitivity and specificity, broad dynamic range for quantification, and the ability to analyze multiple targets simultaneously.
How is real-time PCR used in gene expression studies?
Real-time PCR is widely used to study gene expression by quantifying the amount of mRNA present in a sample. It allows researchers to measure changes in gene expression levels under different conditions or in different tissues.
What are the different fluorescence markers used in real-time PCR?
Commonly used fluorescence markers in real-time PCR include SYBR Green, TaqMan probes, molecular beacons, and hybridization probes. These markers bind to the amplified DNA or RNA and emit fluorescent signals that are measured during the reaction.
How is data analysis performed in real-time PCR?
Data analysis in real-time PCR involves determining the cycle threshold (Ct) value, which represents the cycle number at which the fluorescence signal crosses a predefined threshold. The Ct value is used to quantify the initial amount of the target DNA or RNA in the sample.
Can real-time PCR be used for pathogen detection?
Yes, real-time PCR is commonly used for pathogen detection, including viruses, bacteria, and parasites. It offers high sensitivity and specificity, allowing for the rapid and accurate identification of infectious agents in clinical samples.
What is the difference between absolute and relative quantification in real-time PCR?
Absolute quantification in real-time PCR determines the exact amount of target DNA or RNA present in a sample by comparing the Ct values to a standard curve generated using known concentrations. Relative quantification compares the expression levels of the target gene to a reference gene or control sample without determining the absolute copy number.
Can real-time PCR detect and quantify genetic mutations?
Yes, real-time PCR can be used to detect and quantify genetic mutations by designing specific primers and probes that target the mutated sequence. This allows researchers to identify and quantify the presence of mutant alleles in a sample.