What are Proteins?
- Proteins, often referred to as the building blocks of life, are macromolecules composed of amino acids. These amino acids serve as the foundational units of all proteins. Therefore, to comprehend the nature and function of proteins, one must first delve into the structure and properties of amino acids.
- Amino acids are organic compounds characterized by the presence of both a basic group (-NH2) and an acidic group (-COOH). Central to each amino acid molecule is the α-carbon, to which these groups are attached. Besides these, the α-carbon is also bonded to a hydrogen atom and an organic R group. This R group varies among amino acids, ranging from simple entities like hydrogen atoms to more complex structures. It is this variability in the R group that imparts uniqueness to each amino acid. Interestingly, all amino acids, with the exception of glycine, exhibit chirality due to the asymmetry of the α-carbon. This means they can exist in two optically active forms: D-forms and L-forms. Then, it’s essential to note that amino acids are amphoteric, possessing both acidic and alkaline properties, and typically exist as ions.
- Transitioning from amino acids, proteins are intricate macromolecules made up of one or more chains of these amino acids, connected by peptide bonds. These chains, often referred to as polypeptides, give rise to the diverse range of proteins found in living organisms. The specific sequence of amino acids in a protein determines its structure and, consequently, its function. Therefore, the role of a protein is intrinsically linked to the amino acids it contains.
- Besides their structural role, proteins are pivotal in numerous biological processes. They catalyze metabolic reactions, facilitate DNA replication, respond to stimuli, and transport molecules. Furthermore, proteins provide structural integrity to cells, play a role in cell signaling, and are involved in immune responses. Given their multifaceted roles, proteins are indispensable to the proper functioning of an organism.
- In terms of synthesis, plants have the capability to produce all the amino acids required for protein formation. In contrast, animals lack this comprehensive ability, making certain amino acids essential components of their diet.
- Once synthesized, proteins don’t persist indefinitely. They undergo a process known as protein turnover, where they are degraded and recycled. This turnover ensures that abnormal or misfolded proteins, which could be detrimental to the cell, are swiftly removed.
Definition of Proteins
Proteins are large, complex molecules composed of amino acid chains that perform a vast array of functions in living organisms, including catalyzing metabolic reactions, providing structural support, and facilitating cell signaling and transport.
Properties of Proteins
- Solubility: Proteins typically form colloidal solutions in water rather than true solutions. This behavior can be attributed to the substantial size of protein molecules.
- Molecular Weight: Proteins vary in molecular weight, which is contingent upon the number of amino acid residues they contain. For instance, insulin has a molecular weight of 5,700, while serum albumin weighs around 69,000.
- Shape: Proteins exhibit diverse shapes, ranging from globular forms like insulin to elongated structures as seen in fibrinogen.
- Isoelectric pH: Every protein has an isoelectric pH (pI) determined by its amino acid composition. At this pH, proteins exist as zwitterions or dipolar ions, displaying electrical neutrality and minimal solubility. For example, the pI of pepsin is 1.1, while that of lysozyme is 11.0.
- Acidic and Basic Proteins: The ratio of basic to acidic amino acids in a protein determines its nature. Proteins with a ratio greater than 1 are termed basic, while those with a ratio less than 1 are considered acidic.
- Precipitation of Proteins: Proteins can be precipitated through various methods:
- At pI: Proteins generally exhibit reduced solubility at their isoelectric pH. For instance, casein in milk precipitates at its pI due to lactic acid production.
- By Salting Out: The addition of neutral salts, such as ammonium sulfate, can lead to protein precipitation. This phenomenon, termed salting out, arises from the dehydration of protein molecules.
- By Heavy Metals: Ions like Pb^2+ and Hg^2+ can induce protein precipitation. This principle is employed in using raw egg-white to counteract mercury toxicity.
- By Organic Solvents: Solvents like alcohol can precipitate proteins by dehydrating them. This property underpins the use of surgical spirit as a disinfectant.
- Colour Reactions: Proteins can undergo various color reactions, reflecting the nature of their constituent amino acids. One notable reaction is the Biuret test, where proteins and peptides with at least two peptide linkages produce a purple color upon interaction with dilute copper sulfate in an alkaline medium.
Synthesis of peptides
Peptides are short chains composed of amino acids. These chains, when combined with other peptides, culminate in the formation of proteins, which are vital components of living organisms.
- Formation of Peptide Chains: Peptide chains materialize when two or more amino acids establish a connection through peptide bonds. These bonds are pivotal in holding the amino acids together, thereby giving rise to the peptide structure.
- Variability in Peptide Length: The length of a peptide can vary significantly. A peptide can be as short as comprising only two amino acids. On the other hand, longer chains, known as polypeptides, can encompass between fifty to a hundred amino acids.
- Categorization Based on Length: Peptides are systematically classified based on the number of amino acids they contain:
- Dipeptides: These are peptides that consist of just two amino acids.
- Polypeptides: These are longer chains, containing more than ten amino acids.
- Nature of the Peptide Bond: The bond that forms between amino acids in proteins is a specialized type of amide bond. This bond emerges when the α-carboxyl group of one amino acid molecule fuses with the α-amino group of another amino acid molecule. Therefore, the resulting chain of amino acids, due to the presence of these bonds, is termed a peptide.
- Significance of Peptides: Peptides, being precursors to proteins, play a crucial role in various biological processes. Their synthesis and subsequent combination to form proteins are essential for the proper functioning and structure of living organisms.
Formation of Peptide bond
A peptide bond is a specialized type of amide bond that forms when the α-carboxyl group of one amino acid molecule reacts with the α-amino group of another. This bond is pivotal in linking amino acids together, thereby giving rise to peptides and proteins.
- Condensation Process:
- Initiation: The formation of a peptide bond is initiated when the carboxyl group of one amino acid approaches the amino group of its counterpart.
- Dehydration: As the two groups come closer, a hydroxyl group (OH) is expelled from the carboxyl group, and a hydrogen atom is released from the amino group. Therefore, this results in the elimination of a water molecule (H2O).
- Bond Formation: Subsequently, an amide bond, represented as C-N, is established between the two amino acids, leading to the creation of a dipeptide molecule.
- Nature of Reaction: This entire process is termed a dehydration synthesis reaction, emphasizing the removal of water during protein synthesis.
- Hydrolysis of Peptide Bond:
- Significance: Hydrolysis plays a crucial role in certain synthetic reactions, facilitating the cleavage and transfer of amino acids between peptides.
- Mechanism: During hydrolysis, a water molecule intervenes, breaking the CO-NH bond within the peptide sequence. This action separates the two amino acids, with one retaining the terminal NH2 group and the other possessing the COOH group.
- Catalysis: The presence of acid accelerates the hydrolysis process, ensuring the efficient breakdown of polypeptides into smaller peptides or individual amino acids.
- Understanding Polypeptides:
- Definition: Polypeptides are elongated chains of amino acids, characterized by the presence of more than ten amino acids interconnected by peptide bonds.
- Relation to Proteins: When one or more polypeptides come together, they form a protein, emphasizing the significance of peptide bonds in protein structure.
- Terminology: Each polypeptide has two distinct ends: the amino-terminal (or N-terminal) with a free amino group and the carboxyl-terminal (or C-terminal) with a free carboxyl group.
- Sequence Determination: The sequence of amino acids in a polypeptide is dictated by the codons present in the messenger RNA (mRNA). In turn, these codon sequences in mRNA are derived from the nucleotide sequences in DNA.
Protein Structure
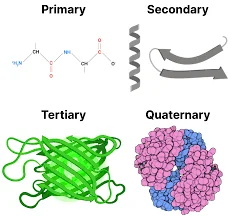
Proteins, the intricate polymers of L-α-amino acids, play a pivotal role in various biological processes. Their structure is multifaceted, and to understand them comprehensively, one must delve into their four levels of organization.
- Primary Structure:
- At its core, the primary structure refers to the linear sequence of amino acids that form the backbone of proteins or polypeptides. This sequence is analogous to the foundation of a building, where amino acids can be likened to individual bricks. The specific order of these amino acids determines the protein’s identity and function.
- Secondary Structure:
- Building upon the primary structure, the secondary structure pertains to the spatial arrangement achieved by the twisting or folding of the polypeptide chain. This structure introduces a new dimension to the protein, much like how a flat wall can be twisted or turned to create architectural nuances.
- Tertiary Structure:
- Delving deeper, the tertiary structure represents the three-dimensional conformation of a functional protein. It is the culmination of various interactions between amino acid residues, leading to a compact and intricate design. In the building analogy, this would be a fully constructed room with all its features and intricacies.
- Quaternary Structure:
- Some proteins transcend the tertiary level and consist of multiple polypeptide chains, known as subunits. The spatial arrangement and interaction of these subunits give rise to the quaternary structure. Imagine a building with various rooms, each distinct yet interconnected, representing this structure.
To draw a parallel, envisioning the structural hierarchy of proteins is akin to understanding the structure of a building. While amino acids are the foundational bricks, the primary structure forms the walls. The twists and turns in these walls represent the secondary structure, leading to a complete room as the tertiary structure. Finally, a building with interconnected rooms symbolizes the quaternary structure.
1. Primary Structure
The primary structure of a protein refers to its most basic form, encompassing one or more linear chains composed of amino acid units. This structure serves as the foundational blueprint for the protein’s overall configuration and function.
- Significance of Amino Acid Sequence: The primary structure distinctly specifies the number and sequence of amino acids that constitute the polypeptide chain. This sequence is pivotal as it determines the protein’s unique characteristics and functionalities.
- Role of Peptide Bonds:
- Formation: Peptide bonds are the primary linkages in proteins, connecting the α-carboxyl group of one amino acid residue to the α-amino group of its neighboring amino acid.
- Composition: Proteins can be composed of single or multiple polypeptide chains, all held together by these peptide bonds.
- Stabilization through Resonance:
- Creation of Partial Double Bond: A partial double bond emerges between the carbon and nitrogen atoms of the amide bond, playing a crucial role in stabilizing the peptide bond.
- Resonance Effect: The nitrogen atom involved in the bond donates its lone pair of electrons to the carbonyl group, initiating a resonance effect.
- Benefits of Resonance: This resonance mechanism not only stabilizes the bond but also restricts the rotation around the C-N bond due to the presence of the partial double bond. The delocalization of electrons across multiple atoms results in a resonance structure that is more stable than the original configuration.
- Configuration and Movement:
- Planar Configuration: The peptide bonds adopt a planar structure, which undergoes minimal movement around the C-N bond.
- Rotational Motion: While the peptide bond itself exhibits limited movement, the single bonds adjacent to the C-N bond display a significant degree of rotational motion, allowing for flexibility in the protein structure.
2. Secondary Structure
The secondary structure of proteins refers to the specific local spatial arrangement of the polypeptide chain. This conformation arises due to the twisting or folding of the chain, where amino acids are positioned close to each other in their sequence. Two primary types of secondary structures have been identified: the α-helix and the β-sheet. The pioneering work of Indian scientist Ramachandran has been instrumental in understanding the spatial arrangement of these polypeptide chains.
- α-Helix:
- The α-helix is a prevalent spiral structure in proteins, characterized by its tightly packed coiled form with amino acid side chains projecting outward from its central axis.
- Proposed by Pauling and Corey in 1951, the α-helix is stabilized by extensive hydrogen bonding between the hydrogen atom attached to the peptide nitrogen and the oxygen atom attached to the peptide carbon.
- Each turn of the α-helix comprises 3.6 amino acids, spanning a distance of 0.54 nm, with each amino acid spaced at 0.15 nm.
- The right-handed α-helix is more prevalent and stable than its left-handed counterpart.
- Certain amino acids, notably proline, and a high concentration of acidic or basic amino acids can disrupt the formation of the α-helix.
- β-Pleated Sheet:
- The β-pleated sheet, another structure proposed by Pauling and Corey, consists of two or more segments of fully extended peptide chains.
- In these sheets, hydrogen bonds form between neighboring segments of the polypeptide chain(s).
- These sheets can be arranged in parallel (same direction) or anti-parallel (opposite direction) configurations.
- β-pleated sheets can be formed by separate polypeptide chains or by a single chain folding back onto itself.
- Many proteins contain β-pleated sheets, often in conjunction with α-helices, forming the core structure of globular proteins.
- Other Secondary Structures:
- Besides the α-helix and β-sheet, proteins also exhibit other secondary structures like β-bends and non-repetitive structures, which are less organized.
3. Tertiary Structure
The tertiary structure of proteins refers to the three-dimensional arrangement of the polypeptide chain. This structure is crucial for the protein’s overall function and stability. In this configuration, the protein adopts a compact form, driven by specific interactions between amino acid side chains.
- Hydrophobic and Hydrophilic Interactions:
- In the tertiary structure, hydrophobic side chains are oriented towards the interior of the protein molecule. This arrangement ensures that these non-polar side chains are shielded from the aqueous environment.
- Conversely, hydrophilic groups, which are attracted to water, are positioned on the protein’s surface. This positioning facilitates interactions with the surrounding aqueous environment and contributes to the protein’s solubility.
- Bonds and Interactions in Tertiary Structure:
- The tertiary structure is not solely maintained by hydrogen bonds. Several other interactions play pivotal roles in stabilizing this structure.
- Disulfide bonds (S-S) are strong covalent bonds formed between the sulfur atoms of two cysteine residues. These bonds provide significant stability to the protein’s structure.
- Ionic or electrostatic interactions occur between oppositely charged side chains of amino acids.
- Hydrophobic interactions arise due to the tendency of non-polar side chains to cluster together, away from water.
- Van der Waals forces, which are weak attractions between molecules, also contribute to the stability of the tertiary structure.
- Domains in Protein Structure:
- A domain is a distinct functional and structural unit within a protein. It represents the basic units of both protein structure (at the tertiary level) and function.
- Typically, a polypeptide chain comprising around 200 amino acids will consist of two or more domains. Each domain can function independently and often has a specific role in the protein’s overall activity.
4. Quaternary Structure
The quaternary structure of proteins refers to the arrangement and interaction of multiple polypeptide chains or subunits within a protein complex. While many proteins consist of a single polypeptide chain, there are several proteins that are composed of multiple chains. These multi-chain proteins exhibit a quaternary structure and are termed oligomers.
- Monomers, Protomers, and Subunits:
- Proteins with a quaternary structure are made up of individual polypeptide chains known as monomers, protomers, or subunits. The terminology used depends on the context, but they all refer to the individual chains that come together to form the complete protein structure.
- A protein consisting of two polypeptide chains is referred to as a dimer, while one with four chains is termed a tetramer.
- Bonds in Quaternary Structure:
- The interactions that hold the individual polypeptide chains together in the quaternary structure are primarily non-covalent in nature.
- These interactions include hydrogen bonds, which are formed between polar side chains; hydrophobic interactions, where non-polar side chains cluster together; and ionic or electrostatic bonds, which occur between oppositely charged side chains.
- Significance of Oligomeric Proteins:
- Oligomeric proteins play a crucial role in various biological processes. They are often involved in the regulation of metabolism and other cellular functions. Their multi-subunit nature allows for intricate regulatory mechanisms and diverse functional capabilities.
- Therefore, understanding the quaternary structure of these proteins provides insights into their functional roles and mechanisms of action.
- Examples of Oligomeric Proteins:
- Hemoglobin is a well-known example of an oligomeric protein. It consists of four polypeptide chains that come together to transport oxygen in the bloodstream.
- Other examples include aspartate transcarbamylase, which plays a role in nucleotide synthesis, and lactate dehydrogenase, an enzyme involved in the conversion of lactate to pyruvate.
Classification of Proteins
Proteins, essential biomolecules with diverse functions, can be classified based on various criteria. The following sections provide a detailed and systematic explanation of the different classifications of proteins:
- Functional Classification of Proteins Proteins can be categorized based on the specific functions they perform in biological systems:
- Structural Proteins: These proteins provide support and structure to various biological entities. Examples include keratin found in hair and nails and collagen present in bones.
- Enzymatic Proteins: These proteins act as catalysts to accelerate chemical reactions. Hexokinase and pepsin are examples of enzymatic proteins.
- Transport Proteins: Proteins like hemoglobin and serum albumin are involved in transporting substances within the body.
- Storage Proteins: Ovalbumin and glutelin are examples of proteins that store nutrients.
- Genetic Proteins: Nucleoproteins play a role in storing and transmitting genetic information.
- Defense Proteins: These proteins, such as snake venoms and immunoglobulins, are involved in defending the organism against threats.
- Receptor Proteins: These proteins bind to specific molecules like hormones and viruses, initiating various cellular responses.
- Chemical Nature and Solubility-Based Classification This classification is based on the amino acid composition, structure, shape, and solubility properties of proteins:
- Simple Proteins: Composed solely of amino acids.
- Subclassification of Simple Proteins
- Globular Proteins: These proteins have a spherical or oval shape, are soluble in water, and are digestible. Examples include albumins, globulins, and glutelins.
- Fibrous Proteins: These proteins have a fibrous structure, are insoluble in water, and are resistant to digestion. Examples include collagens, elastins, and keratins.
- Subclassification of Simple Proteins
- Conjugated Proteins: In addition to amino acids, these proteins contain a non-protein component known as a prosthetic or conjugating group.
- Subclassification of Conjugated Proteins
- Nucleoproteins: These proteins contain nucleic acids as their prosthetic group.
- Glycoproteins: The prosthetic group in these proteins is a carbohydrate.
- Lipoproteins: These proteins are found in combination with lipids.
- Phosphoproteins: The prosthetic group in these proteins is phosphoric acid.
- Chromoproteins: These proteins contain a colored prosthetic group.
- Metalloproteins: These proteins contain metal ions essential for their function.
- Subclassification of Conjugated Proteins
- Derived Proteins: These proteins result from the denaturation or degradation of simple and conjugated proteins.
- Derived Proteins Classification
- Primary Derived Proteins: These are denatured proteins produced by agents such as heat or chemicals.
- Secondary Derived Proteins: These proteins result from the further hydrolysis of primary derived proteins.
- Derived Proteins Classification
- Simple Proteins: Composed solely of amino acids.
- Nutritional Classification of Proteins Based on their nutritional value, proteins can be classified as:
- Complete Proteins: These proteins contain all essential amino acids in the required proportions.
- Partially Incomplete Proteins: These proteins lack one or more essential amino acids but can still support growth.
- Incomplete Proteins: These proteins do not contain one or more essential amino acids and do not support growth.
What is Peptide bond?
In the realm of protein biochemistry, the peptide bond holds a pivotal role. It is the covalent linkage that connects individual amino acids, often likened to bricks, in a protein structure. This bond acts as the cementing material, ensuring the stability and integrity of the protein molecule.
- Formation of the Peptide Bond:
- A peptide bond is established when the amino group of one amino acid reacts with the carboxyl group of another, resulting in the formation depicted in Fig.4.5. It’s essential to note that a dipeptide, which consists of two amino acids, possesses only one peptide bond, not two. When peptides comprise more than ten amino acids, they are termed polypeptides.
- Characteristics of the Peptide Bond:
- The peptide bond is not just a simple linkage; it is planar and rigid, exhibiting characteristics of a partial double bond. Typically, it adopts a trans configuration. The polar nature of both the C O and NH groups in the peptide bond facilitates the formation of hydrogen bonds.
- Representation of Peptide Structures:
- By convention, peptide chains are written with the free amino end, known as the N-terminal residue, on the left, and the free carboxyl end, or the C-terminal residue, on the right. The sequence of amino acids is read from the N-terminal to the C-terminal end, mirroring the process of protein biosynthesis.
- Shorthand for Peptides:
- To simplify the representation of amino acids in peptides or proteins, they are often denoted by a 3-letter or a single-letter abbreviation. This shorthand method streamlines the process of writing proteins. For instance, a tripeptide composed of an N-terminal glutamate, a cysteine, and a C-terminal glycine would be represented as glutamyl-cysteinyl-glycine.
- Naming of Peptides:
- When naming peptides, the suffixes of amino acids, such as -ine, -an, and -ate, are typically transformed to -yl, with the exception of the C-terminal amino acid. Therefore, the tripeptide mentioned earlier, consisting of an N-terminal glutamate, a cysteine, and a C-terminal glycine, is termed glutamyl-cysteinyl-glycine, as illustrated in Fig.4.6.
- Dimensions of a Peptide Chain:
- A fully extended polypeptide chain has specific dimensions, as shown in Fig.4.7. Two neighboring a-carbon atoms are spaced 0.36 nm apart. This figure also highlights the interatomic distances and bond angles, providing a detailed view of the peptide chain’s structure.
Determination of primary structure
The primary structure of a protein is a fundamental aspect that defines its unique sequence of amino acids. This sequence is pivotal for the protein’s function and overall characteristics. Determining the primary structure involves a systematic approach that encompasses several stages.
- Amino Acid Composition:
- The initial step in determining the primary structure is to ascertain the amino acid composition of the protein. This involves complete hydrolysis of the protein to release its constituent amino acids. These amino acids are then quantitatively estimated. Hydrolysis can be achieved through acid or alkali treatment or enzyme hydrolysis. However, enzymatic methods, such as using Pronase—a mixture of non-specific proteolytic enzymes—often result in smaller peptides rather than individual amino acids. Subsequently, chromatographic techniques are employed to separate and quantify the liberated amino acids.
- Degradation into Smaller Fragments:
- Proteins, being large molecules, sometimes consist of multiple polypeptide chains. Therefore, it’s essential to separate these polypeptides before further degradation.
- Liberation of Polypeptides: Agents like urea or guanidine hydrochloride are used to disrupt non-covalent bonds, dissociating the protein into its polypeptide units. To cleave disulfide linkages, performic acid treatment is necessary.
- Number of Polypeptides: The protein’s treatment with dansyl chloride can identify the number of polypeptide chains. This chemical specifically binds with N-terminal amino acids, forming dansyl polypeptides. Upon hydrolysis, these yield N-terminal dansyl amino acids, the count of which equals the number of polypeptide chains in the protein.
- Breakdown of Polypeptides: Polypeptides can be further degraded into smaller peptides either enzymatically, using enzymes like trypsin, chymotrypsin, and pepsin, or chemically using agents like Cyanogen bromide (CNBr).
- Proteins, being large molecules, sometimes consist of multiple polypeptide chains. Therefore, it’s essential to separate these polypeptides before further degradation.
- Amino Acid Sequence Determination:
- Once the polypeptides or their fragments are obtained, the next step is to determine the sequence of amino acids. This process is executed step-by-step to deduce the complete order of amino acids in the protein.
- Sanger’s Reagent: Frederick Sanger employed 1-fluoro 2, 4-dinitrobenzene (FDNB) for determining the structure of insulin. This reagent specifically binds with the N-terminal amino acid, forming a dinitrophenyl (DNP) derivative. Upon hydrolysis, this yields a DNP-amino acid, which can be identified chromatographically.
- Edman’s Reagent: Phenyl isothiocyanate, known as Edman’s reagent, reacts with the N-terminal amino acid to form a derivative. This derivative, when treated with a mild acid, releases a cyclic compound that can be identified chromatographically. The advantage of this reagent is its ability to sequentially degrade a peptide, releasing N-terminal amino acids one by one.
- Sequenator: An advanced method involves using an automatic machine called a sequenator. Based on Edman’s degradation principle, this machine determines the amino acid sequence in a polypeptide. The released amino acid is identified using high-performance liquid chromatography (HPLC).
- Once the polypeptides or their fragments are obtained, the next step is to determine the sequence of amino acids. This process is executed step-by-step to deduce the complete order of amino acids in the protein.
Protein Denaturation
Protein denaturation refers to the process by which a protein loses its native structure. This structural disorganization can lead to the loss of the protein’s secondary, tertiary, and quaternary structures. As a result, the protein undergoes significant changes in its physical, chemical, and biological properties.
- Understanding Protein Denaturation Protein denaturation is the phenomenon where the native structure of a protein becomes disorganized. This disorganization results in the protein losing its specific shape and, consequently, its function. The protein’s secondary, tertiary, and quaternary structures are primarily affected during this process.
- Consequences of Denaturation When a protein is denatured, it undergoes changes in various properties. These include alterations in its physical attributes, chemical reactivity, and biological functions. Therefore, a denatured protein may not perform its intended role within a biological system.
- Agents Causing Denaturation Various agents can induce protein denaturation. These agents disrupt the intricate bonds and interactions that maintain the protein’s structure. They can be broadly categorized into physical and chemical agents:
- Physical Agents:
- Heat: Elevated temperatures can break the hydrogen bonds and other interactions within a protein, leading to denaturation.
- X-rays and UV radiation: These forms of radiation can disrupt the protein’s structure by breaking specific bonds or causing mutations.
- Chemical Agents:
- Acids and Alkalies: These substances can alter the pH, affecting the protein’s charge distribution and leading to structural changes.
- Organic Solvents: Compounds like ether and alcohol can disrupt the hydrophobic interactions within the protein.
- Heavy Metal Salts: Salts of metals like lead (Pb) and mercury (Hg) can bind to specific amino acid residues, altering the protein’s structure.
- Urea: This compound can break hydrogen bonds within the protein.
- Salicylate: This compound can interfere with protein interactions.
- Detergents: Detergents, such as sodium dodecyl sulfate, can disrupt the protein’s lipid interactions, leading to denaturation.
- Physical Agents:
Characteristics of denaturation
Protein denaturation is a complex process that results in the alteration of the protein’s native structure. The following sections detail the specific characteristics associated with denaturation:
- Loss of Native Structure: The protein’s native helical structure is lost during denaturation, leading to a significant change in its conformation.
- Intact Primary Structure: Despite the structural changes, the primary structure of the protein, which consists of peptide linkages, remains intact. This means that peptide bonds are not hydrolyzed during the denaturation process.
- Loss of Biological Activity: A denatured protein loses its biological activity, rendering it non-functional in biological systems.
- Solubility Changes: The denatured protein becomes insoluble in the solvent in which it was originally soluble, leading to precipitation.
- Physical Property Alterations: The viscosity of the denatured protein solution increases, while its surface tension decreases.
- Chemical Changes: Denaturation is associated with an increase in ionizable and sulfhydryl groups within the protein. This is attributed to the loss of hydrogen and disulfide bonds.
- Enhanced Digestibility: Denatured proteins are more easily digested due to the increased exposure of peptide bonds to enzymes. For instance, cooking causes protein denaturation, making cooked food more digestible. Additionally, the denaturation of dietary proteins by gastric HCl enhances their digestion by pepsin.
- Irreversibility: Typically, denaturation is irreversible. An example of this is how an omelet can be made from an egg, but the process cannot be reversed to obtain the original egg.
- Potential for Renaturation: In some cases, careful denaturation can be reversible, a process known as renaturation. For instance, hemoglobin denatured in the presence of salicylate can be renatured upon the removal of salicylate.
- Crystallization: Denatured proteins cannot be crystallized, indicating a loss of their ordered structure.
- Coagulation: Irreversible denaturation can lead to coagulation, where the protein forms a semi-solid viscous precipitate. This coagulation is most effective at the protein’s isoelectric pH. For example, the heat coagulation test is used to detect albumin in urine.
- Flocculation: This refers to the precipitation of proteins at their isoelectric pH, resulting in a substance called flocculum. An example is the precipitation of casein, a milk protein, at its isoelectric pH using dilute acetic acid. While flocculation is reversible, applying heat can convert the flocculum into an irreversible mass known as coagulum.
Biologically Important Peptides
Peptides are compounds consisting of two or more amino acids linked in a chain. In living organisms, several peptides display a broad range of biological functions. Typically, the term “peptide” is used when the number of constituent amino acids is fewer than ten. The following sections provide a detailed overview of biologically active peptides and their respective functions:
- Glutathione:
- Composition: A tripeptide consisting of three amino acids, specifically γ-glutamyl-cysteinyl-glycine.
- Distribution: Found widely in nature in both reduced (2GSH) and oxidized (GSSG) states.
- Functions:
- Serves as a coenzyme for specific enzymes, such as prostaglandin PGE2 synthetase and glyoxylase.
- Prevents the oxidation of sulfhydryl (SH) groups in various proteins, ensuring their proper function.
- Participates in the formation of correct disulfide bonds in proteins.
- Maintains the structure and integrity of RBC membranes.
- Protects hemoglobin from oxidation by agents like H2O2.
- Involved in amino acid transport in the intestine and kidney tubules via the γ-glutamyl cycle.
- Plays a role in detoxification processes, converting toxic substances to mercapturic acids.
- Scavenges toxic peroxides and free radicals through the action of glutathione peroxidase.
- Thyrotropin-Releasing Hormone (TRH):
- Composition: A tripeptide secreted by the hypothalamus.
- Function: Stimulates the pituitary gland to release thyrotropic hormone.
- Oxytocin:
- Composition: A nonapeptide secreted by the posterior pituitary gland.
- Function: Induces contraction of the uterus.
- Vasopressin (Antidiuretic Hormone, ADH):
- Composition: A nonapeptide produced by the posterior pituitary gland.
- Function: Stimulates the kidneys to retain water, thereby increasing blood pressure.
- Angiotensins:
- Composition: Angiotensin I is a decapeptide, which is converted to the octapeptide angiotensin II.
- Function: Angiotensin II has a hypertensive effect and stimulates aldosterone release from the adrenal gland.
- Methionine Enkephalin:
- Composition: A pentapeptide found in the brain.
- Function: Acts similarly to opiates, inhibiting pain sensation.
- Bradykinin and Kallidin:
- Composition: Nonapeptide and decapeptide, respectively.
- Function: Serve as potent vasodilators and are produced from plasma proteins by snake venom enzymes.
- Peptide Antibiotics:
- Examples: Gramicidin, bacitracin, tyrocidin, and actinomycin.
- Function: Exhibit antibiotic properties.
- Aspartame:
- Composition: A dipeptide produced from aspartic acid and phenylalanine.
- Function: Used as a low-calorie artificial sweetener in the soft drink industry due to its sweetness, which is approximately 200 times that of sucrose.
- Gastrointestinal Hormones:
- Examples: Gastrin and secretin.
- Function: Serve as hormones in the gastrointestinal system.
Bonds responsible for protein structure
Proteins, the fundamental building blocks of life, owe their intricate structures to a combination of covalent and non-covalent bonds. These bonds ensure the stability and functionality of proteins. Here is a detailed explanation of these bonds:
- Covalent Bonds:
- Peptide Bonds: These are the primary bonds that link amino acids together in a protein chain. The formation of peptide bonds is a crucial step in protein synthesis.
- Disulfide Bonds: Formed between the sulfhydryl groups (SH) of two cysteine residues, resulting in the formation of cystine. These bonds can be present within a single polypeptide chain or between different polypeptide chains. They play a pivotal role in maintaining the structural integrity and stability of proteins.
- Non-covalent Bonds:
- Hydrogen Bonds: These are formed due to the sharing of hydrogen atoms between the nitrogen and carbonyl oxygen of different peptide bonds. Individually, a hydrogen bond might be weak, but when present in large numbers, they provide significant structural stability to proteins.
- Hydrophobic Bonds: In proteins, the non-polar side chains of neutral amino acids tend to associate closely with each other. While not true bonds in the strictest sense, these hydrophobic interactions are observed in an aqueous environment, causing the molecules to cluster together.
- Electrostatic Bonds: These arise from interactions between negatively charged groups, such as COO– from acidic amino acids, and positively charged groups, like NH3+ from basic amino acids. These interactions contribute to the overall stability of the protein structure.
- Van der Waals Forces: These are subtle forces that occur between electrically neutral molecules. They result from electrostatic interactions due to either permanent or induced dipoles.
Functions of Proteins
Proteins, as fundamental biomolecules, play a myriad of roles in biological systems. Their diverse structures enable them to execute a wide range of functions that are vital for the sustenance and functioning of life.
- Catalytic Activities of Proteins:
- Function: Many proteins serve as catalysts, accelerating the rate of chemical reactions.
- Significance: By enhancing reaction rates, proteins ensure the timely progression of various metabolic pathways, which are essential for the survival and growth of organisms.
- Structural Role of Fibrous Proteins:
- Example: Collagen is a quintessential fibrous protein.
- Function: It acts as a structural unit of connective tissues, providing strength and support.
- Importance: Such proteins are integral in holding skeletal elements together, ensuring the structural integrity of organisms.
- Nucleoproteins and Genetic Inheritance:
- Role: Nucleoproteins are carriers of genetic information.
- Significance: They govern the inheritance of traits, ensuring the transfer of genetic characteristics from one generation to the next.
- Transport Functions of Proteins:
- Function: Proteins regulate the transport of various compounds across cell membranes.
- Importance: This regulation allows cells to maintain internal concentrations of certain compounds, which might otherwise be determined solely by diffusion.
- Regulatory Role of Protein Hormones:
- Function: Protein hormones regulate growth in plants and animals.
- Additional Roles: Besides growth, they also control various physiological functions, ensuring the proper functioning and development of organisms.
- Proteins in Blood Plasma:
- Function: Blood plasma contains soluble proteins beneficial for medical treatments.
- Application: These proteins can be utilized to treat shock resulting from severe injuries or surgical procedures.
- Interferons: The Protective Proteins:
- Origin: Produced by many eukaryotic cells.
- Trigger: Their production is often a response to virus infections, endotoxins, antigenic stimuli, or parasitic organisms.
- Role: Interferons serve as regulatory glycoproteins, offering protection against potential threats.
- Antibiotic Nature of Human Peptides:
- Example: Defensins are peptides derived from humans.
- Function: They exhibit antibiotic properties, providing defense against harmful microorganisms.
References
- Biochemistry, 6e by Satyanarayana (Author)
- Lehninger Principles of Biochemistry Sixth Edition by David L. Nelson (Author), Michael M. Cox (Author)
- https://biologydictionary.net/protein-structure/
- https://lubrizolcdmo.com/technical-briefs/protein-structure/
- https://aklectures.com/lecture/proteins/structure-of-proteins
- https://byjus.com/chemistry/protein-definition/
- https://www.toppr.com/guides/biology/biomolecules/proteins/
- https://www.geeksforgeeks.org/protein-structure/
- https://en.wikipedia.org/wiki/Protein#Structure