What is Polymerase chain reaction (PCR)?
- The Polymerase Chain Reaction (PCR) is a laboratory method that is used to amplify a specific DNA segment many times from very small starting amount.
- The reaction is carried out in vitro, meaning in test tube / reaction vessel, rather than in living cell.
- The technique is based on repeated cycles of three main steps: denaturation, annealing, extension.
- The denaturation step is carried out by heating the DNA to around 90-95 °C so that the double strands separate.
- The annealing step is carried out at a lower temperature so that primers (short synthetic DNA fragments) bind to flanking regions of the target DNA.
- The extension (elongation) step is carried out by a DNA polymerase enzyme which adds nucleotides to the primers to synthesise new complementary DNA strands.
- The polymerase used is thermostable (for example Taq polymerase), so that it remains active despite repeated high temperature denaturation steps.
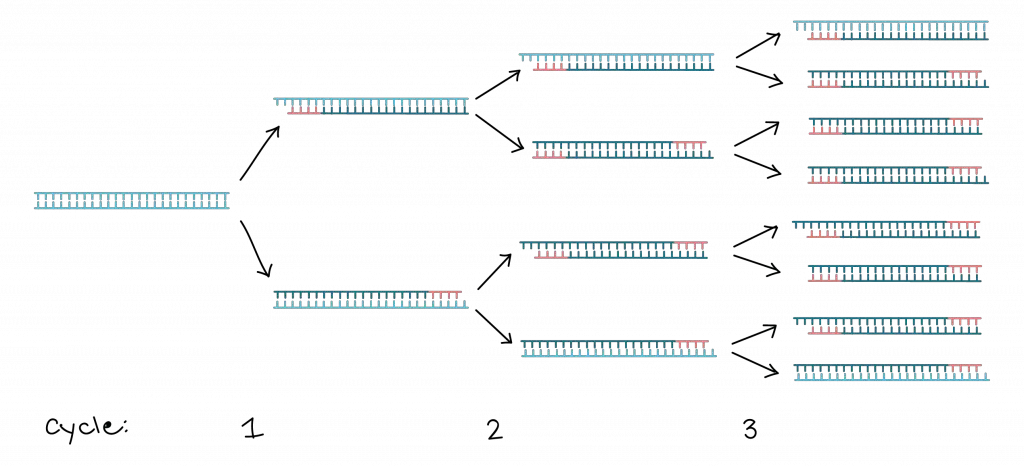
- Millions to billions of copies of the target DNA region are produced after many cycles, because each cycle doubles (approximately) the amount of target DNA.
- The technique is relied upon for many applications: genetic testing, diagnosis of infectious disease, forensic DNA fingerprinting, research in molecular biology.
- Some limitations are present: primer specificity must be ensured, contamination risk is high, and knowledge of target sequence flanking regions is required.
Definition of Polymerase chain reaction (PCR)
Polymerase chain reaction (PCR) is a widely used enzymatic process that rapidly and exponentially amplifies a specific region of DNA, producing millions to billions of copies of a particular DNA sequence.
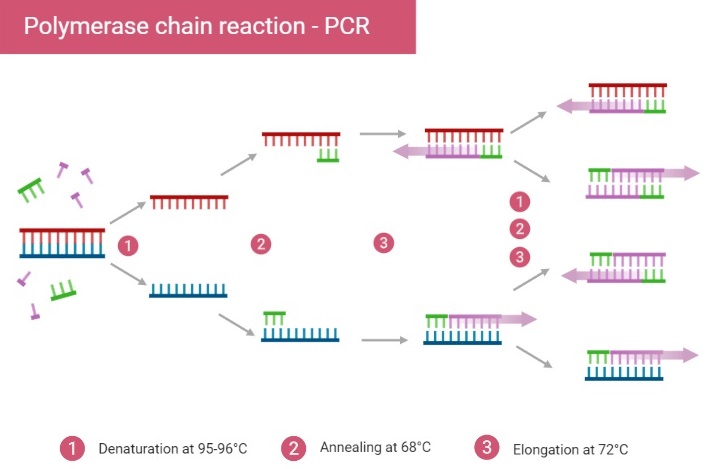
Principle of Polymerase chain reaction
The Principle of Polymerase Chain Reaction (PCR) is based on repeated, controlled cycles of DNA denaturation, primer annealing, and extension, so that a specific DNA segment is amplified exponentially.
Nucleic acid hybridization principle is employed, in which primers (short oligonucleotides) are allowed to bind (anneal) to their complementary sequences on single-stranded DNA templates after denaturation.
Enzymatic replication is used: DNA polymerase adds deoxyribonucleotide triphosphates (dNTPs) to the 3’ end of the primers to synthesise new DNA strands complementary to the template.
Thermal cycling is essential, because temperature changes permit the three main steps (denaturation, annealing, extension) to occur sequentially, which allows amplification to proceed over many cycles.
The template DNA (double-stranded) is first heated to high temperature (≈ 90-95 °C) to break hydrogen bonds and separate into single strands
During annealing, temperature is lowered (commonly ≈ 50-65 °C depending primer melting temperature) so that primers can hybridize to complementary regions on each single strand.
Extension is performed at an optimal temperature for the DNA polymerase (for example ~ 72 °C for Taq polymerase), where the enzyme synthesises new strand by extending from primer based on template strand.
Exponential amplification results, because newly synthesised DNA strands serve as templates in subsequent cycles, so that target region copies grow roughly doubling every cycle (in ideal case) leading to millions-to-billions fold increase.
Specificity is ensured by selection of primers that flank the region of interest; mispriming or non-specific annealing can reduce accuracy.
The DNA polymerase must be thermostable, so that it survives the high temperature denaturation steps; otherwise enzyme would denature and amplification would fail.12
Requirements for PCR
The Components of PCR are required in precise proportion, and every element of mixture plays a critical role, although optimization is often necessary, since too high or too low concentration can disturb amplification efficiency.
- Template DNA –
- The Template is the nucleic acid that contains the target sequence, and it is separated initially into two strands during denaturation.
- DNA may be taken from human blood, saliva, hair, microbes, or any other biological source.
- Purity of the template must be maintained, otherwise inhibitors are carried over which disrupt the reaction.
- The amount depends upon template type, like 1–1000 ng for genomic DNA, but for plasmid or viral DNA, only 1–1000 pg is often sufficient.
- For a standard 50 µl PCR reaction, usually 100–250 ng of mammalian genomic DNA is recommended, while 20 ng of linearized plasmid DNA may be enough.
- Primers –
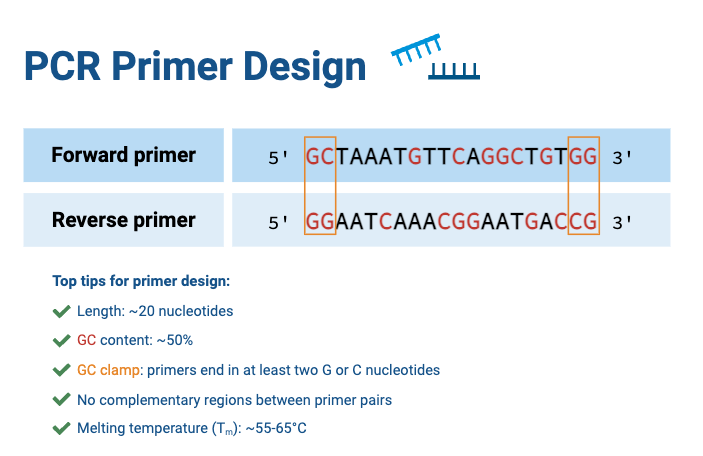
- Primers are short synthetic DNA oligonucleotides, generally 18–40 nucleotides in length, which complement flanking regions of the template.
- A forward primer binds to one strand, and a reverse primer binds to the opposite strand, so both directions of synthesis are initiated.
- The melting temperatures (Tₘ) of primer pair should be within ~5 °C of each other, ideally around 56–62 °C, and 50 % GC content is generally recommended.
- Mismatches at the 3′ end of primer are detrimental, so correct base pairing at this site is critical.
- Too high concentration of primers promotes primer-dimers and non-specific binding, but too low concentration leads to poor amplification, thus usually 0.05–0.1 µM is used.
- Deoxynucleoside Triphosphates (dNTPs) –
- The four dNTPs (dATP, dTTP, dGTP, dCTP) provide building blocks for DNA strand synthesis.
- Typical reactions include ~200 µM of each, although 20–200 µM ranges are used depending on application.
- Higher dNTP concentration increases yield, but excess amounts chelate Mg²⁺ and reduce enzyme fidelity.
- Lower dNTP concentration improves fidelity but decreases overall product yield.
- DNA Polymerase –
- A thermostable enzyme is required, since repeated heating is part of PCR cycles.
- The enzyme extends primers by adding dNTPs to the 3′ hydroxyl end in a 5′→3′ direction.
- Taq DNA polymerase from Thermus aquaticus is most common, but Pfu polymerase is used where high fidelity is demanded.
- Typical usage is ~0.5–2 units per 50 µl reaction, and 1.25 units is considered ideal in most routine amplifications.
- The enzyme’s origin from thermophilic bacteria prevents it from denaturation at high temperatures (90–95 °C).
- Cofactors and Buffer –
- Magnesium ion (Mg²⁺) is the main cofactor, which helps polymerase in forming phosphodiester bonds.
- Optimal Mg²⁺ concentration is around 0.5–5 mM, but it is influenced by template complexity, dNTP concentration, and enzyme type.
- Too much Mg²⁺ increases yield but reduces specificity and fidelity, while too little Mg²⁺ prevents efficient amplification.
- Buffers contain Tris-HCl to maintain pH (~8.4 at room temperature), and KCl or other salts (35–100 mM) to stabilize hybridization.
- For long PCR products, sometimes K+ concentration must be reduced.
Without these components balanced in proper manner, the amplification reaction may not proceed, or products may be non-specific, or yield will be too low.21
Preparation of PCR Reaction Mixture
Add these following ingredients to a PCR tube;
- Sterile Water
- 10X Assay Buffer
- 10 mM dNTP Mix
- Template DNA (100 ng/µl)
- Forward Primer (100 ng/µl)
- Reverse Primer (100 ng/µl)
- Taq DNA Polymerase (3 U/µl)
Note: Tap the tube for 1–2 seconds to mix the contents thoroughly. Then, add 25 μl of mineral oil in the tube to avoid evaporation of the contents. Place the tube in the thermocycler block and set the program to get DNA amplification.
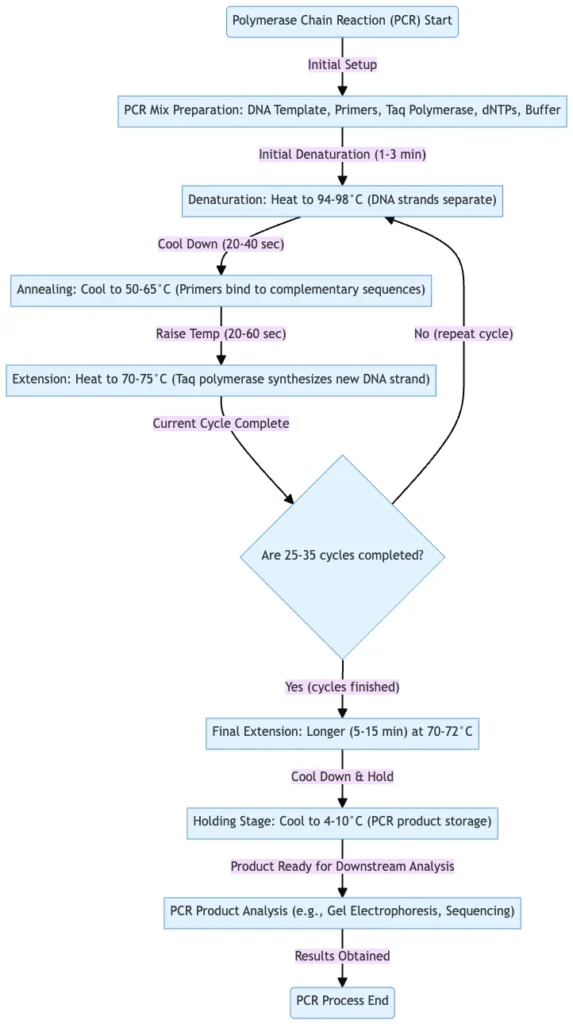
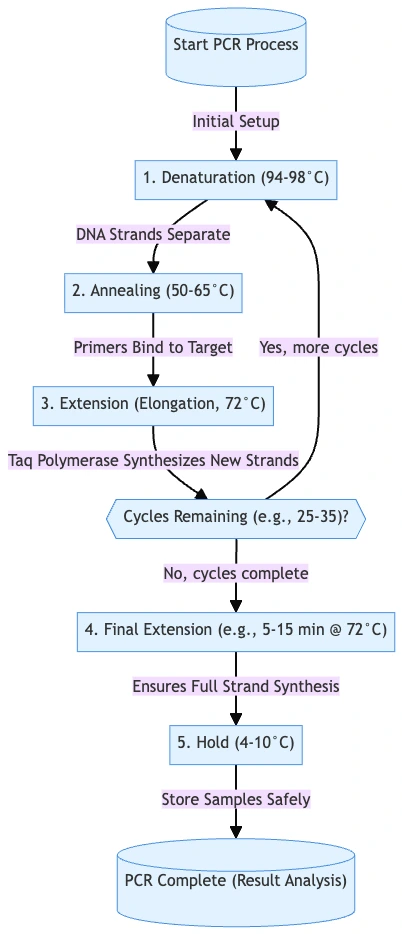
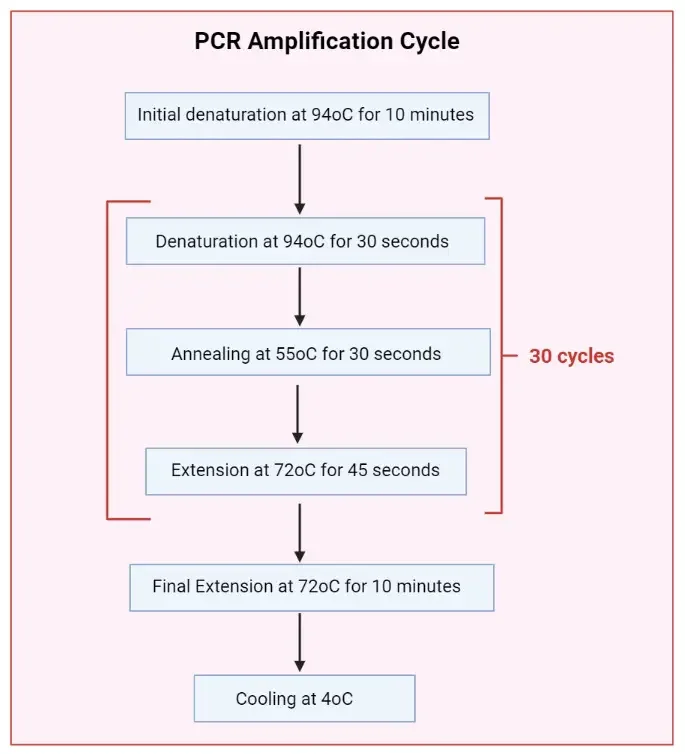
Steps of Polymerase Chain Reaction (PCR)
The Steps of Polymerase Chain Reaction (PCR) are performed in a sequential and also repetitive manner, and each stage is dependent upon the precise condition of temperature and reagents.
- Pre-Preparation Step –
- In this initial step, preparation of PCR reaction mixture is carried out before amplification begins inside thermocycler.
- The DNA or RNA is extracted from biological sample, or sometimes it is taken directly from stored pre-extracted nucleic acid.
- Cleaning of PCR preparation area is performed, and all reagents are equilibrated to working temperature.
- The PCR mixture is made with template DNA, primers, dNTPs, buffer, and enzyme.
- A thermocycler is programmed according to desired protocol, and the reaction mixture is loaded.
- Amplification Step –
- This stage is considered the main process, and it occurs in a cyclic manner, with each cycle having three distinct phases.
- The number of cycles may vary from 20 to 40, depending on required yield.
- Denaturation –
- The double stranded DNA (dsDNA) is heated to about 90–95 °C for 30–90 seconds.
- At this high temperature, hydrogen bonds between the complementary bases are disrupted, and separation into single stranded DNA (ssDNA) occurs.
- The DNA strands are now available to serve as templates.
- Annealing –
- The reaction mixture is cooled down to a lower temperature, generally 55–70 °C, so that primers can hybridize to complementary regions of the ssDNA.
- The forward primer attaches to antisense strand, while reverse primer binds to the sense strand.
- Proper binding of primers provides the starting point for DNA synthesis.
- Elongation / Extension –
- The temperature is adjusted to approximately 72 °C, which is optimum for activity of Taq DNA polymerase.
- The polymerase adds nucleotides to the 3′ end of annealed primers, synthesizing new DNA strand in 5′ → 3′ direction.
- Elongation time depends upon template length, usually 1 kb is synthesized per 0.5–1 minute.
- At the end, two double stranded DNA molecules are produced from one starting molecule.
- Product Analysis Phase –
- After the thermocycling is completed, confirmation of amplification is performed.
- Agarose gel electrophoresis is often employed to visualize amplified DNA fragments.
- In some advanced variations like real-time PCR, product analysis is integrated and performed simultaneously with amplification.56
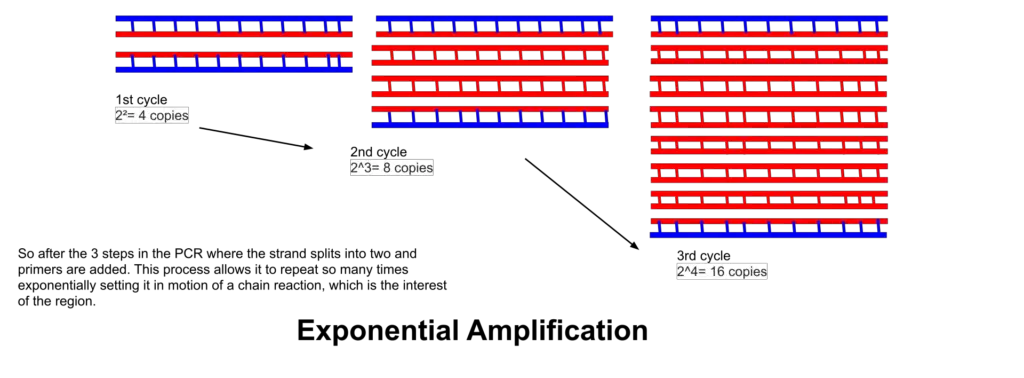
Therefore, by repeating these steps in cycles, exponential amplification of target DNA is achieved, which transforms even a trace amount of nucleic acid into detectable and analyzable quantity.
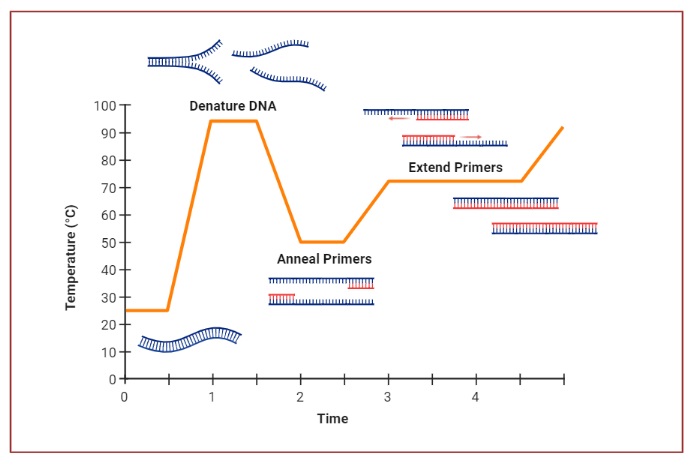
Stages of PCR
The Stages of PCR are divided into distinct phases which are characterized by reaction progress which is observable in many protocols.
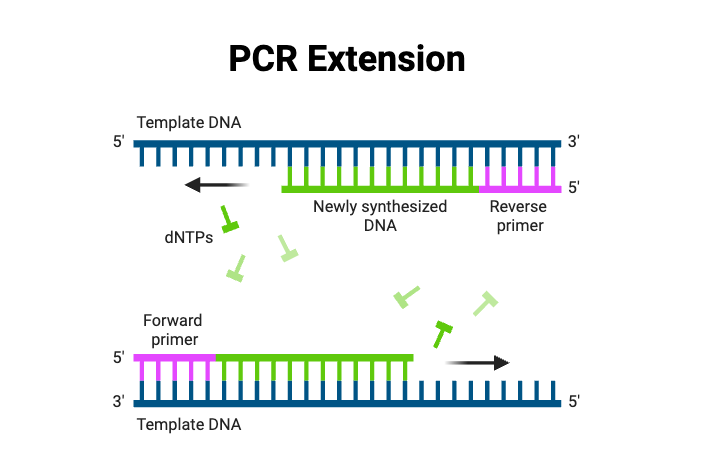
- Exponential Phase ‐
- During early cycles, the amplification of target DNA is nearly ideal, because reagents (primers, dNTPs, polymerase) are abundant, and enzyme fidelity/stability is high.
- Each cycle approximately doubles the number of DNA copies.
- The amount of product is relatively small but increasing rapidly.
- Linear Phase (or Limiting Phase / Leveling Off Stage) ‐
- Reagent depletion (such as dNTPs, primers) begins to affect the reaction, so that doubling is no longer perfect.
- Enzyme efficiency may decline (thermal damage, partial inactivation) which results in slower increment of DNA product.
- Plateau Phase ‐
- Further cycles no longer result in significant increase of product, because one or more essential components has been exhausted, or enzyme has lost activity.
- Background reactions, mis-priming or by-products may accumulate, reducing specificity.
- Final Extensions & Hold Stage ‐
- After last cycling, a final extension step is often employed at ~72 °C to allow any incomplete strands to finish extension.
- A hold at low temperature (eg 4-10 °C) is maintained afterwards, for short-term storage / stabilization of PCR product.
Thus, in summary, PCR performance is influenced by these stages, which affect yield, specificity, accuracy, error rate, etc. The understanding of stages is essential for optimizing PCR protocols.43
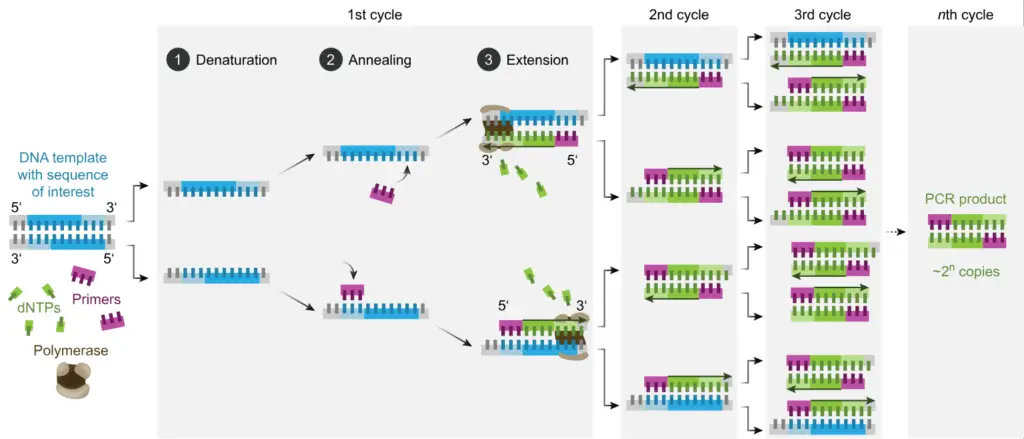
Types of PCR
The Types of PCR have been developed in many variations, and each type was designed for a very specific requirement or application in molecular biology.
- Conventional PCR – It is considered the standard or basic form of PCR, where amplification is performed followed by product analysis using gel electrophoresis.
- Real-Time PCR – Amplification and detection are performed simultaneously in real-time, by using fluorescent dyes or probes. Quantitative results are generated, so it is often used in diagnostics and gene expression studies.
- Reverse Transcriptase PCR (RT-PCR) – RNA molecules are first converted into complementary DNA (cDNA) by reverse transcriptase enzyme. The cDNA is then amplified using normal PCR, allowing detection of RNA templates.
- Quantitative Real-Time PCR (qPCR) – Quantification of nucleic acids is done by monitoring amplification in real-time. Standard curves or comparative Ct methods are used for measuring expression levels.
- Nested PCR – Two successive PCR reactions are carried out using two sets of primers, where the second set binds inside the first amplicon. It increases specificity and sensitivity, but also risk of contamination.
- Multiplex PCR – Several primer pairs are included in a single PCR reaction, so multiple target sequences are amplified simultaneously. It saves time and reagents, but optimization is complex.
- Hot Start PCR – Components are modified or added in such a way that reaction is prevented until high temperature is reached. It reduces non-specific amplification and primer-dimer formation.
- High Fidelity PCR – Special DNA polymerases with proofreading activity (3′→5′ exonuclease) are used. This type is chosen when accuracy of sequence is very important, like cloning or mutational analysis.
- Touch Down PCR – Annealing temperature is gradually decreased in successive cycles. It enhances specificity of primer binding and minimizes non-specific amplification.
- Gradient PCR – It is performed by using a thermocycler with gradient block, so several annealing temperatures can be tested in one run. This type is mainly used for optimization of primers.
- Colony PCR – A bacterial colony is directly used as the template, without prior DNA purification. It is commonly employed in molecular cloning for screening of recombinant colonies.
- Digital PCR – Reaction is partitioned into thousands of micro-chambers or droplets. It allows absolute quantification of nucleic acids, with very high sensitivity.
- Droplet PCR – Similar to digital PCR, but amplification is carried out in water-in-oil emulsion droplets. It is considered highly precise and useful for detecting rare mutations.
- Inverse PCR – Primers are designed in reverse orientation, so unknown flanking regions of DNA can be amplified. It is employed for genomic mapping and transposon identification.
- In Situ PCR – Amplification is performed directly inside cells or tissue sections. It allows localization of nucleic acid sequences within cellular structures.
- Asymmetric PCR – Unequal primer concentrations are used, so one strand is preferentially amplified. It is mainly used for generating single-stranded DNA probes.
- Allelic Specific PCR – Primers are designed to detect single-nucleotide polymorphisms (SNPs). Amplification occurs only if allele-specific primer perfectly matches the template.
- Methylation Specific PCR – DNA is treated with bisulfite, which converts unmethylated cytosines to uracil. Primers distinguish between methylated and unmethylated sequences.
- Repetitive Sequence-Based PCR (rep-PCR) – Primers bind to naturally repetitive DNA elements, so unique banding patterns are produced. It is applied in microbial genotyping and strain differentiation.
- Long Range PCR – Special enzymes or enzyme blends are used to amplify very large DNA fragments, often more than 10 kb. Fidelity and processivity are required for success.
- Overlap Extension PCR (OE-PCR) – DNA fragments with overlapping sequences are joined together by amplification. It is widely used in mutagenesis and gene fusion experiments.
- Linear-After-The-Exponential PCR (LATE-PCR) – Modified asymmetric PCR with improved kinetics, resulting in efficient production of single-stranded DNA.
- Thermal Asymmetric Interlaced PCR (TAIL-PCR) – Used for isolation of unknown sequences adjacent to known sequences. It employs nested specific primers with arbitrary degenerate primers.
- COLD PCR (Co-amplification at Lower Denaturation temperature) – Enrichment of minority alleles or rare mutations is achieved by selective amplification.
- High-Resolution Melt PCR (HRM-PCR) – Following amplification, products are gradually melted while fluorescence is monitored. Differences in melt curves reveal sequence variations like SNPs or methylation.
- Fast Cycling PCR – Special reagents and thermocyclers are used to shorten cycle times. Results are obtained in very rapid manner, sometimes within minutes.
- Alu PCR – Primers are designed for Alu repetitive elements in human genome. It is used for genetic mapping and identification.
- Assemble PCR – Overlapping oligonucleotides are used to build an entirely new DNA sequence. It is frequently applied in synthetic biology.
- Nanoparticle Assisted PCR – Nanoparticles are added into reaction mixture, improving heat conductivity and reaction efficiency.
- Mini Prime PCR – Very small primers are used to generate multiple random amplicons.
- RNase H Dependent PCR – A specially engineered DNA polymerase requiring RNase H activity is employed. It enhances specificity by blocking non-specific primer extension.
- Immuno PCR – Antibody-based detection is combined with PCR amplification of DNA reporter. It allows extremely sensitive detection of proteins.
- In Silico PCR – Computational simulation of PCR is carried out using known genome sequences. It is used for virtual primer testing.
- Suicide PCR – Primers are designed so that they are used only once, avoiding contamination from previous reactions.
- Single Cell PCR – DNA or RNA from a single cell is amplified, enabling single-cell genetic analysis.
- Single Specific Primer PCR (SSP-PCR) – Amplification is carried out using a single primer, often for allele detection.
- Solid Phase PCR – Amplification occurs when primers are immobilized on a solid support like beads or surface. It is commonly applied in next-generation sequencing.
- Variable Number of Tandem Repeats PCR (VNTR-PCR) – Primers flank tandem repeat loci, and variation in fragment length is analyzed. It is widely used in forensic identification and population genetics.
- Intra-sequence Specific PCR – Designed for amplification of sequences within repetitive regions.
From all of these variations, it can be observed that PCR technology has been adapted, modified, and reinvented many times, so that different scientific, forensic, medical, and diagnostic applications can be fulfilled.22
Advantages of PCR
- The Sensitivity of PCR is considered very high, and even a very tiny amount of DNA or RNA is detected because amplification is done in exponential cycles, and even one copy can be multiplied into millions.
- In many cases the Specificity is achieved by primer binding, and reaction is guided toward only target sequence, but sometimes errors may be there if primers bind on wrong sites.
- From the point of view of Speed, results are obtained very rapidly, often within few hours, which is much quicker than some other older laboratory techniques, which sometimes require days.
- The Versatility of PCR has been widely recognized, because it is applied in diagnostics, forensic studies, molecular biology research, and also in environmental monitoring, so many fields has been served.
- Quantitative information is also obtained in some forms, like in Real-Time PCR / qPCR, where not only presence is confirmed, but also expression levels or viral load are measured, giving additional data which is valuable.
- Multiplexing is possible, where multiple primer pairs are included in same reaction tube, so that several target genes are amplified at once, although optimization sometimes is very complicated.
- With automation and high-throughput machines, large number of samples are processed simultaneously, and standardization allows reproducibility of results, although occasional variation is observed.
- A minimal sample requirement is sufficient, and amplification is still successful with degraded or low concentration DNA, therefore less biological material is wasted, which is often an advantage when precious samples are used.
- By using PCR, contamination or infection sources can be identified quickly, and multiple pathogens / genetic variants are detected even in mixed or complex samples.181920
Limitations of PCR
- Prior sequence information is Needed, so primers cannot be designed without knowing flanking regions of target DNA / RNA, which limits detection of novel / unknown sequences.
- Because extreme Sensitivity is Inherent, even minimal contamination of reagents or environment leads to false positives or ambiguous results.
- Inhibition by sample content is experienced, since substances co-purified (eg haemoglobin, phenols, EDTA, humic acids) interfere with polymerase / primer annealing or extension, reducing sensitivity or causing failure.
- Errors during amplification are introduced, because DNA polymerases have imperfect fidelity; base substitutions, insertions/deletions (indels) may occur, misleading sequence interpretation.
- Cost & Complexity are involved, because high-quality reagents, well calibrated thermal cycler, skilled personnel / strict laboratory practices are required for high-throughput or diagnostic PCR.
- Non-specific amplification / Primer-dimer formation is possible, because primers may bind imperfectly or to unintended sequences, reducing yield or generating non-useful products.
- Quantitative accuracy is compromised, because efficiency may vary cycle-to-cycle, plateau effects may appear, or inhibition may distort measurement.
- Amplification of very long templates is difficult, because enzyme processivity / error rate / reaction conditions limit achievable fragment length.14151617
Applications of Polymerase Chain Reaction (PCR)
Applications of PCR are many, and they are used in diverse fields, because amplification of DNA/RNA allows detection / analysis even when starting amount is small
- In medical Diagnostics / Clinical Tests
- Pathogens (viruses, bacteria) are detected rapidly using PCR, especially those which are difficult to culture.
- Genetic disease mutations are screened; carrier status is determined; prenatal diagnosis is done.
- Cancer-related gene expression / oncogene / tumour suppressor mutations are monitored, so treatment decisions can be informed.
- In Research / Molecular Biology
- Gene expression studies are performed by reverse transcription PCR or quantitative PCR to see which gene is turned on/off or how much transcript is present.
- Mutagenesis and cloning are enabled, because specific fragments are amplified and inserted/manipulated for functional studies.
- Sequencing of amplified fragments is done to determine unknown DNA sequences or confirm mutation presence.
- In Forensic / Legal / Identity-Related Work
- DNA fingerprinting is used for crime scene investigation, matching suspects, paternity testing.
- Small or degraded samples (hair, blood stains, ancient remains) are analyzed, because PCR works from small starting material.
- In Agricultural / Environmental / Food Safety Contexts
- Detection of GMOs (genetically modified organisms) is done by PCR, for trait verification or regulatory compliance.
- Monitoring of pathogens / contaminants in water, soil, plants, etc is performed using PCR for early warning or environmental surveys.
- In Quantitative / Real-Time Applications
- Viral load measurement is carried out, and gene expression quantification is enabled via qPCR / RT-qPCR.
- Copy Number Variations (CNVs) are detected more precisely using digital PCR or similar methods, for example rare mutation detection.
- In Evolutionary / Taxonomic / Phylogenetic Studies
- Genetic variation among populations or species is assessed; phylogenetic trees are constructed using PCR of specific marker genes (e.g. rRNA genes).
- Ancient DNA / Archeological samples are studied (e.g. from remains or fossils) using PCR to amplify fragments, for evolutionary or anthropological inference.
- In Forensic / Biosurveillance / Public Health Emergencies
- Outbreak surveillance is supported by rapid PCR tests for emerging pathogens (eg SARS-CoV-2).
- Drug resistance genes in pathogens are monitored; strain typing is performed.
- Others
- Methylation analysis (epigenetics) is done by methylation-specific PCR to see which cytosines are methylated/unmethylated.
- Genotyping / allele / SNP detection is done via allele-specific PCR etc, for research or diagnostic purpose.10111213
Factors affecting Amplification
Many factors affect Amplification in PCR, which must be optimized in order that good yield, specificity, accuracy are obtained.
- Template DNA Quality / Quantity ‐
- Impurities or degraded template DNA cause poor amplification or failure, because polymerase or primers may be inhibited.
- Too much template can lead to non-specific products, or inhibit reaction; too little template yields low product.
- Primer Design and Concentration ‐
- Incorrect primer sequence (mismatches, secondary structures, hairpins) reduces binding specificity, leads to mis-priming / primer dimers.
- Primer melting temperatures (Tₘ) must be compatible (within few °C), otherwise annealing is inefficient.
- Concentration of primers must be balanced; high primer concentration may cause non-specific amplification, low may cause insufficient target amplification.
- Magnesium Ion (Mg²⁺) Concentration / Cofactors ‐
- Mg²⁺ is required by DNA polymerase as cofactor; its concentration influences enzyme activity, primer/template binding, fidelity.
- Excess Mg²⁺ may cause non-specific binding, reduce specificity; insufficient Mg²⁺ may reduce yield or cause failure.
- dNTP Concentration ‐
- Balanced and adequate supply of deoxynucleoside triphosphates is required so that polymerase can extend new strands efficiently.
- Excessive dNTPs may bind cofactors or shift balance, reducing fidelity or causing errors.
- DNA Polymerase Choice / Enzyme Activity ‐
- Thermostability, fidelity, processivity of the polymerase influence amplification, especially for long targets or for high-accuracy requirements.
- Enzyme lot, purity, residual inhibitors may affect performance.
- Annealing Temperature & Time ‐
- Annealing temperature must be optimized in regard to primer Tₘ, GC content, and complexity of template; wrong temperature leads to non-specific binding or no binding.
- Time for annealing must be sufficient but not excessive, because long times at non-optimum temperature may promote mispriming.
- Denaturation Conditions (Temperature & Duration) ‐
- Complete denaturation of template DNA is needed so that strands separate fully; incomplete denaturation reduces primer access.
- Too long or too high temperature can damage enzyme or degrade DNA.
- Extension Time & Temperature ‐
- Extension temperature must match enzyme’s optimum; usually around 72 °C for many polymerases.
- Extension time must be sufficient for target length; longer amplicons need more time.
- Cycle Number ‐
- Too few cycles produce low yield; too many cycles may lead to plateau effect, non-specific products, increased errors.
- Template GC Content / Secondary Structure ‐
- High GC content causes stronger base-pairing, complicates denaturation and primer binding; secondary structures (hairpins etc) can impede primer annealing or extension.
- Inhibitors / Contaminants ‐
- Substances co-extracted with template (e.g. phenols, heme, soil chemicals) may inhibit polymerase or interfere with binding.
- Carry-over of reaction by-products, salts etc may reduce efficiency if not removed or diluted.
- Buffer Composition & Salt Concentration ‐
- Buffers provide required pH, ionic strength; wrong buffer composition reduces enzyme activity or changes primer binding.
- Salt concentration (e.g. Na+, K+) influence annealing specificity and melting temperatures.789
FAQ
What is PCR?
PCR is a laboratory technique used to amplify a specific DNA sequence, creating millions or billions of copies of the target DNA. It is based on the principles of DNA replication and involves a cyclic process of denaturation, annealing, and extension using DNA polymerase.
What is the purpose of PCR?
The primary purpose of PCR is to amplify a specific DNA sequence of interest. It is widely used in research, diagnostics, forensic analysis, genetic testing, and various other applications where the detection and analysis of DNA are necessary.
How does PCR work?
PCR involves repeated cycles of temperature changes to facilitate DNA denaturation, primer annealing, and DNA synthesis. The DNA sample is subjected to cycles of heating to separate the double-stranded DNA, cooling to allow primers to bind to the target sequence, and DNA synthesis by a heat-stable DNA polymerase.
What are the key components needed for PCR?
The essential components of a PCR reaction include a DNA template containing the target sequence, DNA primers that flank the target sequence, DNA polymerase (such as Taq polymerase), nucleotides (dNTPs), buffer solution, and magnesium ions (Mg2+).
What are the different types of PCR?
Some common types of PCR include conventional PCR, real-time PCR (qPCR), multiplex PCR, nested PCR, reverse transcription PCR (RT-PCR), and quantitative PCR (qPCR). Each type has specific variations and applications.
What is the role of primers in PCR?
Primers are short DNA sequences that bind to complementary regions flanking the target DNA sequence. They serve as starting points for DNA synthesis by the DNA polymerase. Primers are critical for target specificity and amplification.
What is the significance of PCR in genetic testing?
PCR is widely used in genetic testing to identify genetic mutations, detect infectious agents like viruses and bacteria, diagnose genetic diseases, determine gene expression levels, and perform DNA profiling for forensic analysis and paternity testing.
How can PCR be used in research?
PCR plays a crucial role in various research applications, including cloning, DNA sequencing, gene expression analysis, DNA fingerprinting, mutation detection, genotyping, and studying microbial diversity and evolution.
What are the limitations of PCR?
PCR has some limitations, such as the possibility of amplifying nonspecific DNA sequences if the primers are not specific enough, the potential for contamination leading to false results, the requirement for prior knowledge of the target sequence, and difficulties in amplifying highly GC-rich or repetitive DNA regions.
What is the role of PCR in the COVID-19 pandemic?
PCR has been instrumental in the diagnosis of COVID-19 by detecting the presence of SARS-CoV-2 viral RNA in patient samples. It has been widely used for mass testing, tracking the spread of the virus, and monitoring the effectiveness of vaccination campaigns.
