Native PAGE is the method where proteins are separated in their original folded condition without any use of denaturing reagents. It is the process in which the movement of proteins in a polyacrylamide gel is controlled by the natural charge, size and the native conformation of the molecules.
The proteins migrate through the gel matrix as it acts like a molecular sieve. In this technique the non-covalent interactions among the subunits are not disturbed, so the protein complexes remain intact during the separation.
This is referred to as an important system for studying oligomeric state, protein–protein interaction and enzymatic activity because the proteins is obtained in active form after electrophoresis. It is important that the samples are not heated during preparation as heating may disturb the native structure.
Some of the main variations of this method are Clear Native PAGE (CN-PAGE) and Blue Native PAGE (BN-PAGE), in which the separation of high-molecular-weight complexes is performed with better clarity.
Principle of Native Polyacrylamide Gel Electrophoresis (PAGE)
The principle of Native PAGE is based on the separation of proteins in their natural folded condition so the structural features and biological activity are preserved. It is the process where the movement of proteins depends on their net electrical charge, molecular size and the native conformation present in solution.
The polyacrylamide gel works like a molecular sieve and the proteins migrate according to their charge density, which is the net charge in relation to their mass or surface area. In alkaline running buffer the proteins carrying higher negative charge move faster towards the anode while the protein complexes having larger mass face more resistance during movement. It is the gel matrix that creates frictional force and this force slows down the bigger molecules while the smaller ones pass through the pores easily. Since no denaturing agent is added, the non-covalent subunit interaction is maintained and the protein remains in active state.
This is referred to as an important condition for studying oligomeric form, protein–protein interaction and enzymatic activity, and the whole separation pattern is controlled completely by the natural charge to mass ratio of the proteins.
Types of Native Polyacrylamide Gel Electrophoresis
Native PAGE can be performed in different forms and each type is designed to maintain the folded structure of proteins while changing the way charge or buffer condition influence the movement. It is the process in which separation is controlled either by the intrinsic charge of the protein or by adding specific dyes or mild detergents that keep the native conformation intact. Some of the main types are as follows–
1. Standard/Traditional Native PAGE– It is the common method where proteins are separated based on their natural charge, size and conformation. No anionic dye is used and the migration depends only on the native charge to mass ratio of the proteins present in the sample.
2. Blue Native PAGE (BN-PAGE)– This is referred to as a high-resolution method because Coomassie Brilliant Blue G-250 binds non-covalently to the proteins giving a uniform negative charge. The separation is now controlled mainly by the molecular size and hydrodynamic shape. It is helpful for estimating molecular weight and studying stoichiometry of large complexes.
3. Clear Native PAGE (CN-PAGE)– It is the mildest technique and is performed without any dye. The movement of the protein depends completely on the intrinsic net charge and the pore size of the gradient gel. It is used for in-gel enzymatic assays where Coomassie dye may interfere and it also helps in maintaining fragile and labile complexes.
4. High-Resolution Clear Native PAGE (hrCN-PAGE)– It is a modification of CN-PAGE where mixed micelles like sodium deoxycholate and DDM are used in the cathode buffer. It is designed to increase the resolution while keeping the proteins in native condition.
5. Native SDS-PAGE (NSDS-PAGE)– This method uses very low SDS concentration and avoids heating. It is the process where the resolution of SDS-PAGE is obtained but the protein still retains its native cofactors and enzymatic activity because the denaturation is minimal.
6. Native Isoelectric Focusing (Native IEF)– It separates proteins according to their isoelectric point in a pH gradient while the structure of proteins remains unchanged. It is often used as the first dimension in 2D electrophoresis.
7. Tris–Glycine System– It follows the Laemmli system and works in alkaline pH. It is useful for studying proteins in the range of 20–500 kDa where the native charge is maintained during migration.
8. Tris–Acetate System– This system runs closer to neutral pH and is recommended for very large proteins above 150 kDa. It maintains the protein stability better compared to high-pH systems.
9. NativePAGE Bis-Tris System– It works at around pH 7.5 and is mainly used for membrane and hydrophobic proteins. When combined with Coomassie G-250 it behaves similarly to BN-PAGE and separates proteins primarily according to their molecular weight.

Requirements for Polyacrylamide Gel Electrophoresis (PAGE)
Some of the main requirements are–
- Gel Matrix (Polyacrylamide)
It is the medium used for separation. It is formed by acrylamide and bis-acrylamide polymerization. APS and TEMED is used for initiating polymer formation. The pore size depend on the percentage of acrylamide used, and it is the process that allow proteins of different sizes to move at different rates. - Buffer System
A discontinuous buffer system is used. It is the process where different buffers are taken in electrode chambers and gel preparation. These buffers maintain pH and provide ions for current flow. - Electrophoresis Apparatus
A vertical gel tank, gel cassette and a power supply is required. The gel is placed between the anode and cathode so that the proteins can move in the electric field. - Operational Conditions
It is usually run at constant voltage. PAGE may be run for long duration depending on gel percentage and the type of proteins.
Sample Preparation Requirements
A. Requirements for Native PAGE
In this method protein structure must be preserved. It is the process where no denaturing agent should be used.
Some important requirements are–
- The samples is prepared without SDS, reducing agent or heating.
- The sample buffer contain sucrose or glycerol for increasing density so that the sample sink properly.
- Proteins must be in soluble form. Aggregated proteins give distorted bands.
- Genomic DNA must be removed because it increases viscosity.
- High salt concentration must be avoided as it cause streaking of bands.
- Membrane proteins may require mild detergents for solubilisation.
B. Requirements for SDS-PAGE
This is the denaturing form where proteins are separated only by molecular weight.
- SDS must be present in gel, sample buffer and running buffer.
- Reducing agents like DTT or β-mercaptoethanol is added for breaking disulfide bonds.
- The sample is heated (70–85°C) so that complete unfolding occur.
Buffer Systems for PAGE
Some important buffer systems are–
- Tris-Glycine Buffer
It is used in many PAGE methods. The pH remain alkaline. It is the process suited for a wide range of proteins. - Tris-Acetate Buffer
It is used when very large proteins move slowly. The near neutral pH help in maintaining stability. - Bis-Tris System (NativePAGE / BN-PAGE)
It is near-neutral pH system. A cathode buffer additive like Coomassie G-250 is used. This dye gives uniform negative charge to proteins. - Clear Native PAGE (CN-PAGE)
This technique does not use dyes. The protein move according to their intrinsic charge. Mild detergents may be used when membrane proteins are analysed.
Molecular Markers
Protein markers of known molecular mass is required. It helps in calculating the size of unknown proteins after separation.
Procedure of Native Polyacrylamide Gel Electrophoresis
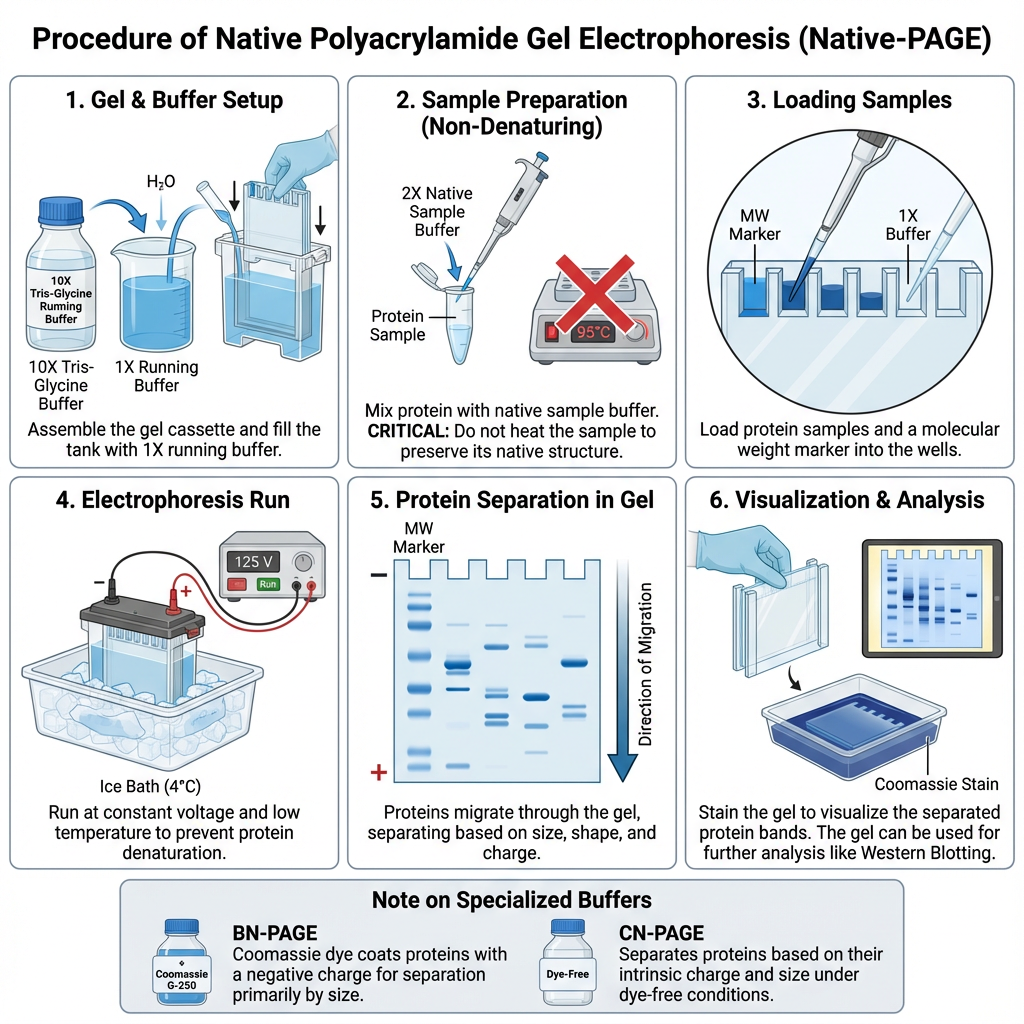
1. Preparation of running buffer and gel setup
- It is the process where the 10X Tris-Glycine Native Running Buffer is diluted to 1X.
- If a discontinuous Bis-Tris system is used in BN or CN PAGE then the anode and cathode buffers is prepared separately.
- The electrophoresis cassette is taken from the pouch, rinsed and placed in the chamber with the notched side facing the cathode tank.
- The comb is removed gently and the wells is rinsed with 1X running buffer to clear any bubbles or debris.
- After this the upper (inner) and lower (outer) chambers is filled with appropriate 1X running buffer.
2. Specialized buffer preparation (BN-PAGE / CN-PAGE)
- For BN-PAGE the cathode buffer is prepared by adding Coomassie Brilliant Blue G-250 (or SERVA Blue G) to a final concentration of about 0.002% (w/v).
- This is used to give a net negative charge to the proteins.
- In CN-PAGE the buffer does not contain any anionic dye and the system runs under dye-free native conditions.
3. Sample preparation (non-denaturing)
- The protein sample is mixed with equal volume of 2x Native Sample Buffer which normally contains 10% glycerol and a tracking dye (Bromophenol Blue).
- The sample must not be heated because native structure and activity is preserved in this method.
- The concentration of sample is checked so that the load remains around 10 µg per well and overloading is avoided.
- Empty wells is filled with 1X sample buffer.
4. Loading and electrophoresis
- The prepared samples and native molecular weight markers (21–720 kDa range) is loaded into the wells without exceeding almost 3/4th well capacity.
- The chamber is closed and connected to the power supply.
- Wells that do not contain sample is filled with 1X sample buffer to keep conductivity uniform.
5. Running conditions
- The run is carried out at constant voltage and at low temperature (ice bath or 4 °C) for preventing denaturation and aggregation.
- In Tris-Glycine Native PAGE the gel usually runs at 125 V constant for 1–12 hours.
- In Bis-Tris systems a two-step protocol is used where 50 V is applied for about 10 minutes followed by 200 V for nearly 120 minutes.
6. Post-electrophoresis analysis
- After completion the power is switched off and the cassette is opened carefully.
- The gel is removed and processed for detection.
- The proteins is visualized by fixing and staining the gel with Coomassie dyes such as SERVA Blue R or Coomassie Brilliant Blue G-250.
- In some cases the gel is used for in-gel activity assays (zymography) where deposition of chromophore or precipitate confirms activity.
- BN/CN-PAGE can also be used as first dimension in 2-D electrophoresis or transferred to PVDF membrane for Western blotting.
SDS PAGE vs. Native PAGE
| Aspect | SDS PAGE | Native PAGE |
|---|---|---|
| Gel Nature | Denatured gel | Non-denatured gel |
| Denaturation | SDS is used to denature proteins | No denaturation step required |
| Separation Principle | Based on protein mass | Based on protein size and charge |
| Protein Stability | Proteins are not stable and cannot be recovered | Proteins are stable and can be recovered |
| Protein Conformation | Proteins are unfolded | Proteins retain their native conformation |
| Protein Activity | Protein activity is lost | Protein activity is preserved |
| Sample Preparation | Requires boiling in SDS sample buffer | Minimal or no sample preparation required |
| Resolution | Higher resolution due to denaturation | Lower resolution due to native conformation |
| Applications | Analysis of protein molecular weight and purity | Analysis of protein complexes and native structures |
Applications of Native Polyacrylamide Gel Electrophoresis (PAGE)
- It is used for studying oligomerization state of proteins such as monomer, dimer, trimer or higher forms.
- It helps in analyzing protein complexes and interactions under native conditions.
- BN-PAGE and CN-PAGE is used for studying OXPHOS complexes and their assembly intermediates.
- It is the method used for checking protein integrity and aggregation before structural determination.
- Detergent screening for membrane proteins is done by BN-PAGE to see which detergent maintain monodispersity.
- It verifies oligomeric state that is observed in crystallization studies.
- It is used for in-gel enzymatic activity assays (zymography) to detect active enzymes inside the gel.
- CN-PAGE is preferred for activity assays where Coomassie dye may interfere.
- It can be used for monitoring enzyme kinetics when imaging systems is used during running.
- It helps in verifying functional integrity of proteins after purification.
- It is used for analyzing charge heterogeneity and isoforms formed by post-translational modifications.
- It helps in detecting conformational changes caused by ligand binding or mutation.
- It is used to compare different native proteins because migration depend on their net charge and shape.
- It is used as first dimension in 2D electrophoresis followed by SDS-PAGE in second dimension.
- Protein bands separated in native gels is excised for further mass spectrometry analysis.
- CN-PAGE is useful for native MS because no dye is present.
- NSDS-PAGE is used in metalloproteomics for retaining metal cofactors of metalloproteins.
Advantages of Native Polyacrylamide Gel Electrophoresis (PAGE)
- It preserves the natural folded state of proteins during separation.
- It allows proteins to retain biological and enzymatic activity.
- Non-covalent interactions is generally maintained which help in studying tertiary and quaternary structure.
- It provides conditions that closely mimic physiological environment.
- It is used for determining oligomerization state of proteins.
- It helps in studying protein–protein interactions and stable complexes.
- It allows in-gel enzymatic assays (zymography) because activity is preserved.
- It can be used for continuous monitoring of enzyme kinetics when imaging systems is applied.
- It helps in detecting charge heterogeneity and conformational changes caused by PTMs or ligand binding.
- BN-PAGE gives high resolution and helps in estimating native molecular mass.
- Coomassie dye in BN-PAGE gives uniform negative charge which improve separation.
- BN-PAGE helps in preventing aggregation of membrane proteins.
- It is used for detergent screening to check monodispersity before crystallization.
- CN-PAGE provide milder conditions and preserve fragile complexes.
- CN-PAGE is useful for dye-sensitive enzymatic assays.
- CN-PAGE is suitable for native MS because no dye removal is required.
- Proteins can be recovered from native gels in active form.
- It is used as first dimension in 2D electrophoresis.
- Neutral pH buffer systems help in stabilizing large protein complexes.
Limitations of Native Polyacrylamide Gel Electrophoresis (PAGE)
- It often gives lower resolution compared to SDS-PAGE.
- CN-PAGE generally shows lower resolution than BN-PAGE.
- Accurate molecular weight estimation is difficult because migration depend on charge, size and shape together.
- CN-PAGE has variable migration since it rely only on intrinsic net charge.
- Smearing and poor band detection is common in native gels.
- Protein aggregation is a major problem especially for hydrophobic membrane proteins.
- DNA contamination in lysates increase viscosity and disturb migration.
- High salt concentration can distort mobility and produce streaking.
- Very alkaline pH in Tris-Glycine system may cause band distortion or loss of resolution.
- Proteins with high pI may migrate towards cathode which complicate interpretation.
- If running pH is close to protein pI then migration becomes very slow or stops.
- Coomassie dye in BN-PAGE can interfere with in-gel activity assays.
- BN-PAGE is not suitable for native MS because dye removal affect protein conformation.
- Nitrocellulose membrane cannot be used after BN-PAGE since it binds dye strongly, so PVDF is required.
- Some fragile complexes may dissociate during BN-PAGE conditions.
- Even high-resolution CN-PAGE may fail with membrane proteins because aggregation still occur.
Agarose vs polyacrylamide gel electrophoresis
| Aspect | Agarose Gel Electrophoresis | Polyacrylamide Gel Electrophoresis |
|---|---|---|
| Composition | Natural polysaccharide extracted from seaweed. | Synthetic polymer formed from acrylamide monomers. |
| Pore Size | Larger pores suitable for separating large DNA fragments. | Smaller pores ideal for resolving proteins and small nucleic acids. |
| Applications | Commonly used for DNA and RNA analysis, including fragment sizing and purification. | Primarily used for protein analysis and small nucleic acid fragments, such as in SDS-PAGE. |
| Resolution | Lower resolution; less effective for distinguishing molecules with small size differences. | Higher resolution; capable of separating molecules with minor size variations. |
| Preparation | Easier to prepare; gels are typically cast horizontally and can be remelted and reused. | Requires careful handling due to toxicity; gels are usually cast vertically and cannot be reused. |
| Toxicity | Considered non-toxic and safer to handle. | Acrylamide is a neurotoxin; proper protective measures are necessary during preparation and handling. |
| Cost | Generally less expensive, making it suitable for routine analyses. | More costly due to the complexity of preparation and materials involved. |
FAQ
What is Native Polyacrylamide Gel Electrophoresis (PAGE)?
Native PAGE is an electrophoretic technique used to separate proteins based on their size and charge under non-denaturing conditions. It preserves the native structure and function of proteins, allowing analysis of protein-protein interactions, oligomerization, and complex formation.
How does Native PAGE differ from SDS-PAGE?
Unlike SDS-PAGE, which uses denaturing agents to unfold proteins and separate them solely based on size, Native PAGE maintains the native conformation of proteins, separating them based on both size and charge. It provides information about native protein structures and interactions.
What is the purpose of Native PAGE?
Native PAGE is commonly used to study protein complexes, oligomerization, and protein-protein interactions. It helps in identifying different protein isoforms, determining native molecular weights, and investigating protein assembly/disassembly processes.
What is the gel composition used in Native PAGE?
The gel matrix used in Native PAGE typically consists of polyacrylamide and a non-ionic detergent, such as Triton X-100, to solubilize membrane proteins while preserving their native conformation.
What are the advantages of Native PAGE over other protein separation techniques?
Native PAGE allows the analysis of native protein structures and interactions without denaturation, preserving their biological activity. It can provide valuable insights into protein complexes, protein-protein interactions, and molecular weight estimation under native conditions.
How can Native PAGE be used to determine protein oligomerization?
Native PAGE separates protein complexes based on their size and charge. By comparing the migration patterns of known protein complexes with the migration of individual proteins, one can determine the oligomeric state of the protein of interest.
Can Native PAGE be used for quantification of protein samples?
Quantification of proteins using Native PAGE can be challenging due to various factors, such as variable staining or detection methods. However, relative quantification can be achieved by comparing band intensities within the same gel under standardized conditions.
Can Native PAGE be combined with other techniques for further analysis?
Yes, Native PAGE can be combined with other techniques such as immunoblotting, mass spectrometry, or activity assays to provide additional information about protein identification, post-translational modifications, or functional characterization.
What are the limitations of Native PAGE?
Some limitations of Native PAGE include limited resolution, difficulty in standardizing mobility, lower sensitivity compared to other techniques, and the potential for protein aggregation or loss during electrophoresis.
How can gel-to-gel variability in Native PAGE be minimized?
To minimize gel-to-gel variability, it is important to maintain consistent gel preparation conditions, such as gel composition, pH, buffer conditions, and handling techniques. Proper standardization and the use of appropriate controls can help in reducing variability between experiments.
- Arndt, C., Koristka, S., Bartsch, H., & Bachmann, M. (2012). Native Polyacrylamide Gels. Protein Electrophoresis, 49–53. doi:10.1007/978-1-61779-821-4_5
- https://www.med.unc.edu/pharm/sondeklab/wp-content/uploads/sites/868/2018/10/Native-gel-analysis.pdf
- https://www.thermofisher.com/in/en/home/life-science/protein-biology/protein-gel-electrophoresis/protein-gels/specialized-protein-gels/nativepage-bis-tris-gels.html
- http://www.assay-protocol.com/molecular-biology/electrophoresis/native-page.html
- http://www.assay-protocol.com/molecular-biology/electrophoresis/diverse-native-PAGE.html
- https://gyansanchay.csjmu.ac.in/wp-content/uploads/2022/10/Native-PAGE.pdf
- https://www.bio-rad.com/webroot/web/pdf/lsr/literature/Bulletin_6040.pdf
- https://molbio.mgh.harvard.edu/szostakweb/protocols/native_page/index.html
- https://www.differencebetween.com/difference-between-sds-page-and-vs-native-page/