How L-arabinose operon different from other operons?
- This operon is responsible for the cellular degradation of arabinose compounds.
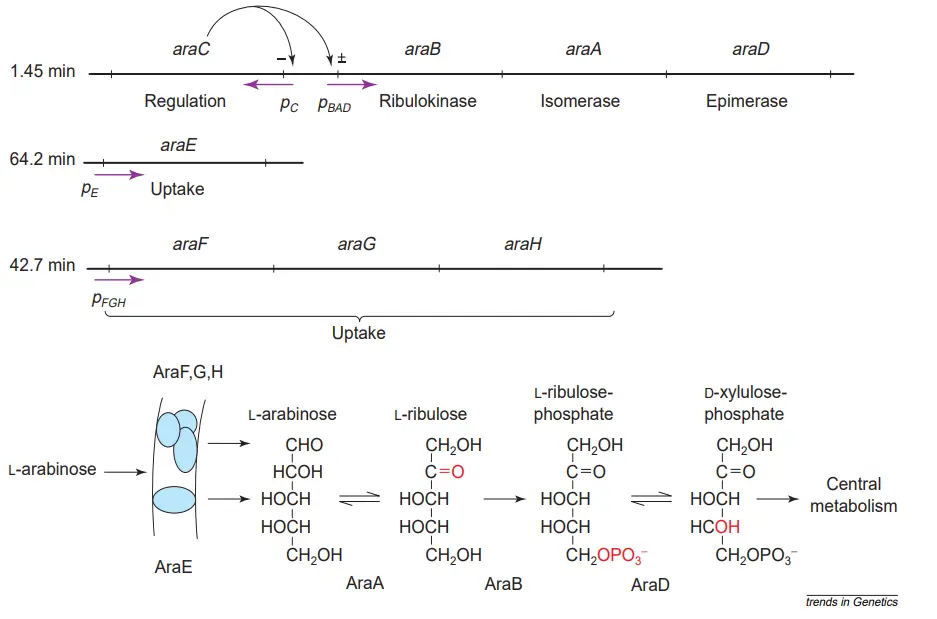
- Arabinose is transformed to ribulose by arabinose isomerase, which is encoded by the araA gene, then phosphorylated by ribulokinase, which is encoded by the araB gene, and finally converted to xylulose-5-phosphate by ribulose-5-phosphate epimerase, which is encoded by the araD gene.
- The final product enters the pentose phosphate pathway and produces reducing power or precursor glycolysis metabolites.
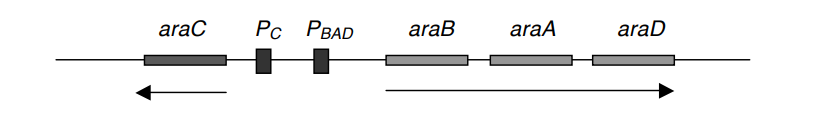
- These three structural genes share a single promoter, PBAD, and are controlled by AraC, the product of the araC gene.
- This operon appears to be identical to the lac operon at first glance. In the absence of arabinose, AraC is generated and linked to araC. Thus, araC serves as its own regulator.
- In the absence of arabinose, the binding of AraC to araC inhibits the attachment of RNA polymerase (RNAp) to PBAD, preventing transcription of all genes.
- When arabinose becomes accessible, however, it binds to AraC and triggers a structural shape shift, rendering PBAD susceptible to RNAp’s attachment.
- The proposed model for this operon is shown below. PC is the promoter of araC.
Nevertheless, Englesberg et al. discovered that the ara operon’s function differs significantly from that of the lac operon.
- In most cases, the lac operon is negatively regulated, but the ara operon can be positively or negatively controlled, depending on the conditions.
- Although lacI mutations result in constitutive lac operon expression, araC mutations do not. In fact, mutations in araC result in a “super-repressed” state in which araA, B, and D are inhibited even in the presence of copious arabinose.
- Constitutive mutants are common in a negatively regulated operon like as the lac operon, but they are exceedingly uncommon in the ara operon. This demonstrated that the constitutive phenotype in the ara operon may not be related to araC inactivation.
Thus, the following paradigm was hypothesised:
- This model implies that two AraC molecules are always associated as a dimer. Furthermore, this dimer can exist in both active (P1) and inactive (P2) forms (P2).
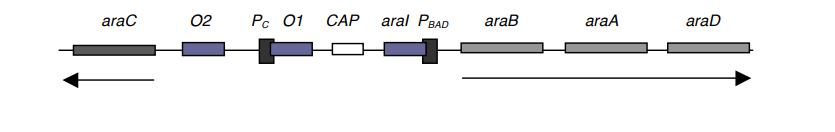
- AraC dimer is in the P1 state when arabinose is absent, but when arabinose is present, it can react with AraC and shift its conformation to P2, allowing it to attach to the araI site of the DNA.
- The location of araI is between PBAD and the CAP site (see diagram). Upon attachment of a P2 AraC to araI, transcription of araB, A, and D occurs.
- Similar to its role in the lac operon, catabolite activator protein is also involved in the regulation of arabinose.
- This model suggests two operators, O1 and O2, which are also AraC binding sites.
- When AraC binds to any of these operators, araA, B, and D transcription is inhibited. Mutations in araC, called araCC, alter AraC such that it remains in the P2 state permanently, causing the operon to remain in the “on” position even in the absence of arabinose.
- It was discovered that only a small number of specific amino acids in AraC transformed it to araCC, which explains why such mutations are so uncommon.
- Overall, AraC is required for the function of the araB, A, and D genes, and if the araC gene is deleted (e.g., by transposon insertion), there will be no transcription of the araB, A, and D genes.
- It should be noted that more recent research (e.g., Johnson & Schleif, 1995, J. Bacteriol. 177:3438-3442) indicates that the ara operon is considerably more complex than previously described, with three distinct processes in the cell regulating arabinose use.
The arabinose system: genes and behavior
- The arabinose system allows E. coli and its relatives to take up pentose L-arabinose from the growth medium utilising products of the unlinked araE and araFGH genes, and then convert intracellular arabinose in three steps catalysed by the products of the araBAD genes to D-xylulose-5-phosphate.
- This is then introduced into the pentose phosphate shunt. The AraC protein regulates the expression of its own synthesis and other ara system genes.
- In the presence of arabinose, AraC stimulates the commencement of mRNA synthesis from the unknown function gene’s promoters pE, pFGH, pBAD, and pJ.
- At pBAD, the AraC protein not only works positively to induce transcription in the presence of arabinose, but also operates negatively to repress transcription initiation in the absence of arabinose; whereas at pC, AraC acts negatively regardless of the presence or absence of arabinose9.
- Utilizing rifamycin-permeable cells, careful tests were conducted using arabinose to induce and rifamycin to inhibit mRNA production. These results indicate that the synthesis of araBAD mRNA begins within three seconds of the administration of arabinose.
- Experiments on the unlinked araE and araFGH genes lack such time resolution, but indicate that these genes require around five minutes to be fully induced.
- AraC induces the protein products of the araBAD genes to approximately 300 times their level without induction. The inducibility of the genes is decreased in the presence of glucose and certain other carbohydrates, a phenomenon referred to as “catabolic sensitivity.”
- This sensitivity is mediated by the quantity of cyclic AMP, which modulates the activity of the cyclic AMP receptor protein, CRP, which, in addition to AraC, is required for the complete stimulation of the arabinose genes6.
- D-fucose (5-methyl-L-arabinose), a structurally similar analogue of L-arabinose, inhibits cell growth on arabinose.
- Fucose binds to AraC but generally does not stimulate transcription. Fucose-induced growth inhibition is a geneticist’s dream, as the isolation of fucose-resistant mutants is extremely straightforward.
- The fucose-resistant mutations are located in AraC, and some of them render the system constitutive, meaning that the mutant AraC protein stimulates transcription even in the absence of arabinose.
- Other mutations are also activated by fucose in addition to arabinose. Fucose-resistant mutants were essential for demonstrating that the ara system is positively regulated.
How the ara system works?
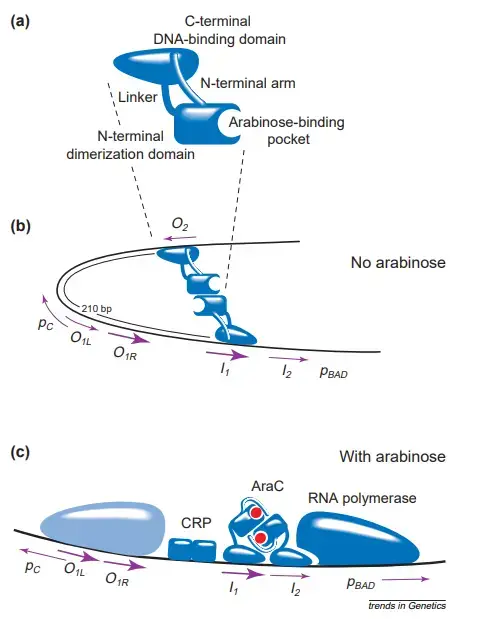
- AraC protein has a homodimeric structure. The monomer possesses two domains: a dimerization domain that binds arabinose as well as a DNA-binding domain.
- These domains are loosely connected by a flexible linker that enables the dimeric protein (in the presence of arabinose) to bind to half-sites in their natural direct-repeat orientation separated by four bases, the natural spacing between the I1 and I2 or O1L and O1R elements, or by an additional 10 or 21 bases (one or two helical turns of the DNA) in artificial constructs.
- Additionally, the flexible linker allows the protein to connect to inverted half-sites.
- In the absence of arabinose, an N-terminal arm consisting of 18 amino acids extends from the dimerization domain and attaches to the ‘back side’ of the DNA-binding domain.
- The combination of the arm and the linker holds each DNA-binding domain to its dimerization domain pretty tightly.
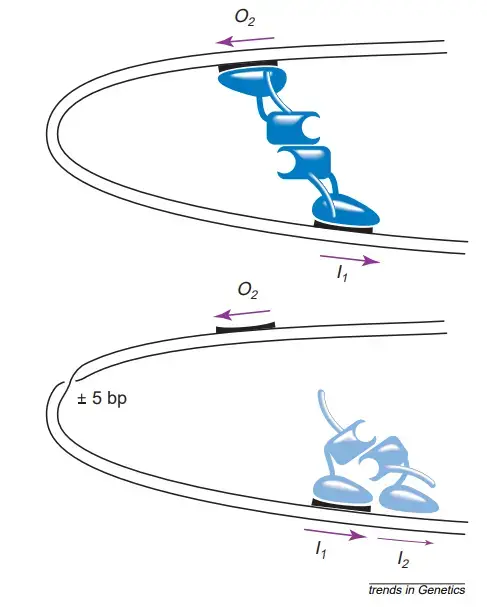
- The DNA-binding domains are properly oriented for binding to the I1 and O2 half-sites and generating a DNA loop, but are utterly misoriented for binding to neighbouring half-sites.
- Binding to adjacent half-sites (i.e., binding cis) would necessitate either a significant bending of AraC or the disruption of at least one of the arm–DNAbinding domain contacts.
- AraC is therefore energetically disfavored to bind to I1–I2 in the absence of arabinose, but energetically favoured to attach to nonadjacent half-sites and construct a DNA loop.
- When arabinose binds to the dimerization domains, it is energetically preferable for the N-terminal arms of AraC to bind to the dimerization domains rather than the DNA-binding domains.
- The arabinose-mediated liberation of the arms from their connections with the DNA-binding domains loosens the restrictions that hold them, allowing the domains to reorient and assume any relative orientation.
- Importantly, the now-flexible AraC makes it possible for the overall energy state of the DNA–AraC system to be lower if AraC binds to the nearby I1 and I2 half-sites as opposed to looping between I1 and O2.
- As an aside, X-ray crystallography reveals that arabinose induces very minute changes in the structure of the core of the dimerization domain; hence, it is likely that the direct interactions between arabinose and the arms themselves are the primary determinant of the positions of the arms.
- The DNA loop that is generated in the absence of arabinose contributes to the arabinose system in numerous ways.
- First, its presence sterically limits RNA polymerase’s access to the pBAD promoter, hence maintaining a low level of pBAD expression.
- In addition to preventing RNA polymerase from attaching to the pC promoter, the loop may also prevent the cyclic AMP receptor protein from joining to its binding site alongside AraC.
- Looping to O2 prevents AraC from occupying the I2 half-site and activating pBAD transcription improperly.
- Having AraC bound to the regulatory region in the absence of arabinose enables it to respond rapidly to the addition of arabinose, as the potentially slow DNA-binding process has already occurred.
- The transition of the arms from the DNA-binding domains to the dimerization domains does not involve controlled movement of the arms or domains, nor does the shift of the DNA-binding domains from looping to binding cis.
- Random diffusion should be sufficient. It samples the arabinose occupancy of the dimerization domain because the arm is in equilibrium and continually dissociates from and reassociates with the two domains.
- In general, the arm spends the majority of its time bound to the DNA-binding domain in the absence of arabinose and the dimerization domain in the presence of arabinose.
- As a result of diffusion, the DNA-binding domain that was bound to the O2 half-site can relocate to the I2 half-site and induce pBAD within a few seconds of arabinose’s arrival.
- The transitory derepression of the pC promoter upon arabinose addition is an unexplained phenomena. Shortly after the administration of arabinose, the activity of this promoter increases approximately tenfold, but returns to its pre-induction level approximately ten minutes later.
- This response is consistent with the concept that the opening of the DNA loop upon the addition of arabinose permits RNA polymerase unrestricted access to the pC promoter until AraC can bind at the O1 site.
- AraC may then bind to O1 slowly due to its low concentration in the cytoplasm, as most AraC may have coupled nonspecifically to random DNA sequences as a result of arabinose addition.
- This concept describes in detail one of the benefits of DNA looping, namely that it facilitates rapid induction kinetics of arabinose catabolism enzymes.
DNA looping: discovery, proof and biological use – helical-twist experiment
- DNA looping was discovered in a textbook-like manner. Englesberg had published results demonstrating that a specific deletion entering the pBAD regulatory area from upstream resulted in the loss of repression by AraC, despite the promoter’s ability to be triggered normally.
- Large-scale genetic mapping of the operon was prompted by the prospect that a negative control element may be upstream of all the parts required for positive control of pBAD.
- Unpredictably, this research revealed that the negative control element, now known as O2, might be located hundreds of base pairs upstream of all the sequences required for positive control.
- The discovery and demonstration of DNA looping proved this point. The DNA looping by AraC between a site within pBAD and a site well upstream explained how a negative regulatory element may work from a position far upstream of a promoter.
- The crucial test of DNA looping was the helical-twist experiment, in which a half turn of the DNA helix was introduced anywhere between the upstream site required for complete suppression of pBAD and the downstream site required for induction.
- Such a half-turn rotates one of the two sites to which AraC binds to the opposite side of the DNA, significantly impeding the production of DNA loops.
- This experiment can show the existence of DNA looping if the binding energies of AraC to the two half-sites are not adequate to twist the DNA and return the half-sites to their original orientation.
- Introducing half-rotations decreased repression of pBAD, whereas introducing integral numbers of rotations did not. This not only offered significant support for the looping theory, but also enabled for the in vivo assessment of DNA’s helical twist.
- Consistent with the conclusion that relatively little energy is available for DNA looping in the ara system was the observation that the upper boundary for DNA looping and trans-binding by AraC in the absence of arabinose is only slightly larger than the natural loop size of 210 base pairs (i.e. 500 base pairs).
- The precise value of the corresponding lower boundary is unknown, however it is fewer than 100 base pairs.
- The fundamental prediction of the DNA looping hypothesis was that AraC would occupy the I site even in the absence of arabinose.
- Prior to this discovery, it was believed that gene regulation was primarily performed by modulating the binding or dissociation of a protein from DNA, and it was unanticipated that AraC would remain bound to a portion of the I site in the absence and presence of arabinose.
- To test this essential hypothesis, in vivo footprinting using dimethyl sulphate was created. This approach is fortunate in that AraC bound to the I1 and more distant O2 half-sites significantly improves the interaction of dimethyl sulphate with guanine in the sites.
- Using in vivo footprinting, it was possible to demonstrate that both the O2 halfsite and the I1 halfsite are occupied in the absence of arabinose, and that arabinose decreases O2 occupancy.
- The fact that the binding of AraC to the O2 half-site is eliminated when the I1 half-site is mutated such that AraC cannot initiate transcription from pBAD is one of the most crucial pieces of supporting evidence from the footprinting.
- As predicted by the DNA-looping theory, the binding of AraC to I1 and O2 involves substantial cooperation.
- Before DNA looping could be replicated in vitro, it was impossible to determine the stoichiometry of AraC associated with looping.
- Experiments have demonstrated that a dimer of AraC interacts to linear DNA at the I1 –I2 site33.
- Consequently, it was likely that DNA looping required four subunits, two coupled to I and two bound to O2. In vitro DNAlooping tests with supercoiled minicircles revealed that the DNA is instead looped by a dimer of AraC.
- Therefore, one monomer interacts with O2 and one monomer interacts with I1. Experiments on DNA looping in vitro demonstrated that DNA looping in the ara system does not occur efficiently until the DNA is supercoiled.
- The available energies for DNA looping are apparently insufficient to create the loop unless the DNA is already partially looped due to the existence of supercoiling strain.
- Obviously, the low energy involved were crucial to the success of the helical-twist tests.
- The low energies are also suitable for a regulatory mechanism, as a 100-fold shift in the equilibrium state of the ara regulatory system takes only a tiny energy input, equivalent to the change in one hydrogen bond in water or two conventional van der Waals interactions.
- Looping was discovered in several other bacterial systems, including lac, deo, gal, and gln, following the discovery of DNA looping in the ara system.
- DNA looping eventually became the recognised explanation for how eukaryotic enhancers might work remotely.
- DNA looping has multiple functions for cells. First, it permits multiple proteins to affect RNA polymerase and the initiation process, some from adjacent sites and others from distal sites, and second, the cooperativity inherent in the use of multiple DNA-binding sites increases the effective binding constants and enables regulatory proteins to function at extremely low concentrations.
- Text Highlighting: Select any text in the post content to highlight it
- Text Annotation: Select text and add comments with annotations
- Comment Management: Edit or delete your own comments
- Highlight Management: Remove your own highlights
How to use: Simply select any text in the post content above, and you'll see annotation options. Login here or create an account to get started.