What is Heterogeneous Nuclear RNA (hnRNA)?
- Eukaryotic mRNAs, unlike prokaryotic mRNAs, are monocistronic. Heterogeneous nuclear RNA describes the primary transcript in eukaryotes, which is substantially larger than mature mRNA (hnRNA).
- It contains sequences that are distinct and has approximately 10 times as many sequences as mature mRNA.
- hnRNA is processed to form mRNA; hence, it is referred to as “mRNA precursor” or “pre-mRNA.”
- The word hnRNA refers to the unprocessed mRNA (pre-mRNA) molecules found in the nucleus.
- It is composed primarily of pre-mRNA molecules, which require intensive processing to mature into mRNA molecules.
- The heterogeneous nuclear ribonucleoprotein is composed of hnRNA linked with proteins (hnRNP).
- The first stage in mRNA synthesis is transcription. RNA polymerase reads the DNA sequence during transcription in order to produce a corresponding, antiparallel RNA strand (called primary transcript).
- Pre-mRNA refers to the original transcript that eventually forms messenger RNA (mRNA).
- The pre-mRNA must then undergo additional processing to produce a mature, functional mRNA.
Properties of hnRNA
- Size is not uniform and sedimentation values range from 20S to 100S.
- Base composition of hnRNA is similar to DNA from which it is transcribed.
- Mostly the hnRNA is located outside the nucleolus.
- mRNA is derived from hnRNA through enzymatic processing.
Processing of hnRNA
During processing, the hnRNA undergoes the following chemical modifications:
1. Packaging of hnRNA with protein
- hnRNAs that have just been produced are combined with protein to form a ribonucleoprotein complex.
- Protein particles consist of 30S particles and are dispersed on hnRNA.
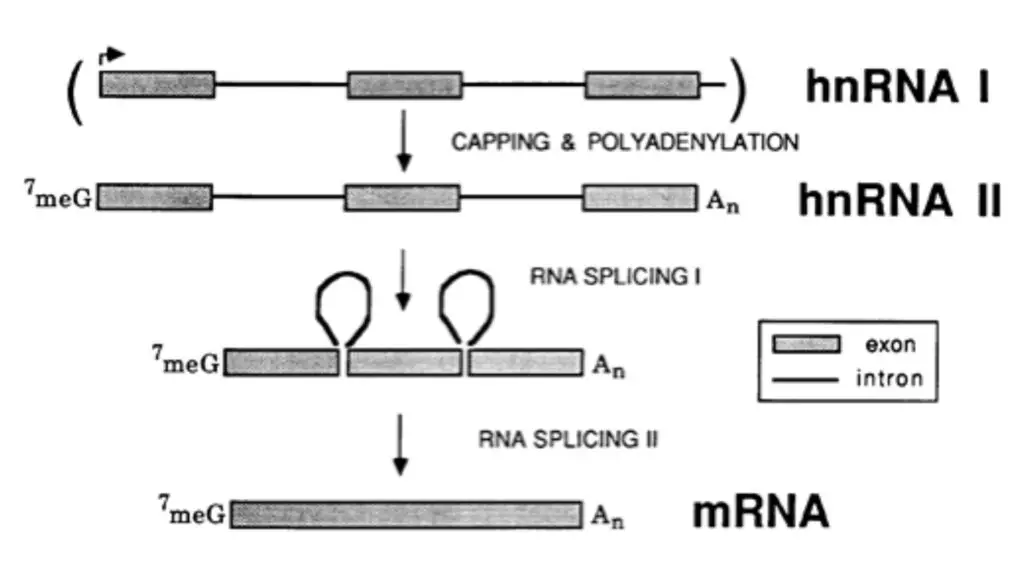
2. Polyadenylation or Poly (A) tail formation
- The enzyme “poly (A) polymerase” catalyses the polymerization reaction following the addition of a poly (A) tail to the 3′-end of the RNA after transcription.
- This enzyme adds approximately 200 adenine nucleotides to the 3′ OH- end of hnRNA without a DNA template.
- The purpose of the Poly (A) tail is to transfer mRNA from the nucleus to the cytoplasm.
- Additionally, it slows the intracellular breakdown of mRNA.
- Poly (A) tail is not present in all eukaryotic mRNAs, such as histone mRNA.
3. RNA splicing
- The hnRNA is composed of coding sequences (exons) and “intervening sequences” (introns).
- These sequences are found alternately; such genes are referred to as split genes.
- Genes vary in the number of exons and introns they include. In chicken, the actin gene comprises four exons and three introns, whereas the collagen gene contains fifty-one exons and fifty introns.
- In extreme circumstances, the actual coding region of mRNA comprises approximately 10% of hnRNA’s length.
- RNA splicing is the process by which introns are removed and exons are joined in the correct order during processing.
- Some “snurp” ribonucleoprotein particles are involved in the removal of RNA introns and RNA splicing.
- Similar base sequences at the end of each intron are referred to as “splice junctions.” The RNA of the “snurp” has sequences complementary to “splice junctions” and is involved in the intron removal process.
- However, not all genes require the snurps for RNA processing. Some mitochondrial and ribosomal RNA genes in ciliated protozoa undergo RNA self-spicing.
4. 5′ Capping
- At the 5′ end of RNA, a “cap” is generated during processing.
- The cap is composed of one or two methylated nucleotides at the 2′ position.
- The methylated nucleotide is coupled to 7-methylguanosine by a triphosphate bridge (m7G).
- 7-methylguanosine is connected with 5′-triphosphate at the 5′-end of mRNA, and as a result, it terminates in the 3′-OH position.
- The terminal modified base (m7G-PPP) of the cap protects the mRNA from 5′-exonuclease assault and destruction.
- In contrast to bacterial mRNAs, eukaryotic mRNAs are long-lived molecules. They can endure for several hours to days. In seeds in dormancy, they can live for years. Transport of mature mRNA to the cytoplasm.
Mature mRNA
One central coding segment, two untranslated segments, one on either side of the central coding segment, 7-methylguanosine linked by a triphosphate linkage at the 5′-end, and a 200-base-long Poly(A) tail at the 3′-OH end make up mature mRNA.
Genes vary in the length of their mRNA, coding region, and untranslated portions. For instance, the human a-globin gene contains 575 bases of total mRNA. The coding region contains 429 bases, but the 5′-end and 3′-end untranslated portions include 37 and 109 bases, respectively.
1164 bases make up the coding region of the ovalbumin mRNA from chicks, whereas 64 bases at the 5′-end and 631 bases at the 3′-end make up the two untranslated segments.
References
- Calvet, J. P., & Pederson, T. (1977). Secondary structure of heterogeneous nuclear RNA: Two classes of double-stranded RNA in native ribonucleoprotein. Proceedings of the National Academy of Sciences, 74(9), 3705–3709. doi:10.1073/pnas.74.9.3705
- Hancock, John. (2004). hnRNA (Heterogeneous Nuclear RNA). 10.1002/0471650129.dob0325.
- Torelli, U. (1975). Characteristics of Heterogeneous Nuclear RNA in Normal Small Lymphocytes and in Acute Leukemia Blast Cells. Acta Haematologica, 54(4), 234–241. doi:10.1159/000208080
- https://www.genscript.com/biology-glossary/1350/heterogeneous-nuclear-RNA
- http://www.med.upenn.edu/dreyfusslab/publications/1984bDreyfussMolCellBiol.pdf
- https://byjus.com/question-answer/explain-the-process-of-making-heterogenous-nuclear-rna-hnrna-into-a-fully-functional-mrna-in/
- https://www.dictionary.com/browse/heterogeneous-nuclear-rna
- https://www.biologyonline.com/dictionary/hnrna
- http://allie.dbcls.jp/pair/hnRNA;heterogeneous+nuclear+RNA.html
- https://www.biologydiscussion.com/cytogenetics/macromolecules/heterogeneous-nuclear-rna-hnrna-protein/35886
- https://www.sciencedirect.com/topics/medicine-and-dentistry/heterogeneous-nuclear-rna