Eukaryotic gene regulation is a fundamental aspect of molecular biology that plays a critical role in the expression of genes within eukaryotic cells. This intricate system governs when, where, and how much specific genes are expressed, which is essential for the proper development and functioning of organisms, ranging from unicellular protozoa to complex multicellular organisms, including humans.
At its core, eukaryotic gene regulation encompasses a series of multi-layered processes that influence various cellular activities, such as metabolism, growth, differentiation, and responses to environmental changes. The regulation of gene expression is not only crucial for normal cellular function but also for the understanding of various diseases. Abnormalities in gene expression are often linked to numerous conditions, including cancer, genetic disorders, and neurological diseases. This highlights the importance of studying eukaryotic gene regulation within the realm of biomedical research, where insights can lead to potential therapeutic strategies.
Unlike prokaryotic cells, eukaryotic cells exhibit a more complex system of gene regulation. One key distinction is the spatial separation of transcription and translation. In eukaryotes, transcription occurs in the nucleus, where the DNA is tightly wrapped in a structure called chromatin. This packaging adds an additional layer of complexity, as the accessibility of DNA to the transcription machinery is tightly regulated. The chromatin can exist in different states, either tightly packed (heterochromatin) or loosely organized (euchromatin), which directly influences gene expression.
Regulation occurs at several levels, including transcriptional, post-transcriptional, translational, and post-translational stages. At the transcriptional level, various transcription factors bind to specific DNA sequences in the promoter regions of genes, either promoting or inhibiting their expression. Enhancers and silencers, which are additional regulatory elements located far from the target gene, can also influence transcription by interacting with the transcription machinery.
Post-transcriptional regulation involves modifications to the RNA transcripts, such as splicing, capping, and polyadenylation. These modifications not only enhance the stability of mRNA but also play a crucial role in determining the types of proteins that are ultimately produced. Furthermore, small RNA molecules, such as microRNAs, can bind to mRNA molecules and regulate their stability and translation, showcasing another layer of complexity in gene regulation.
Translational regulation occurs at the ribosome level, where various factors can modulate the initiation and elongation of protein synthesis. Finally, post-translational modifications, such as phosphorylation, glycosylation, and ubiquitination, can significantly affect a protein’s function, stability, and localization within the cell. These modifications provide a rapid means of regulating protein activity in response to various stimuli.
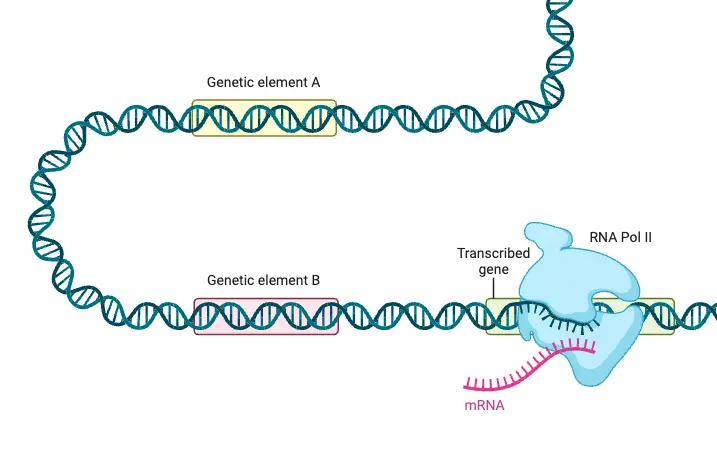
Gene Regulation in Eukaryote
Gene regulation in eukaryotes is a complex and essential process that governs how genes are expressed within cells. Unlike prokaryotes, eukaryotic gene regulation involves multiple layers and mechanisms, reflecting the intricate organization of their genomes, which are housed within a nuclear envelope and associated with chromatin. This complexity allows for precise control over gene expression, crucial for development, growth, and environmental responsiveness.
- Types of Gene Regulation in Eukaryotes:
- Transcriptional Regulation:
- Definition: This process involves controlling gene expression at the transcription level, thereby influencing how efficiently genetic information is transcribed from DNA into messenger RNA (mRNA).
- Mechanisms: It encompasses several elements, including transcription factors that can act as activators or repressors, chromatin remodeling to alter DNA accessibility, and modifications to histone proteins that can affect the packaging of DNA.
- Post-transcriptional Regulation:
- Definition: This type of regulation occurs after the transcription of DNA into mRNA and influences various aspects of mRNA, including its processing, transport, stability, and eventual translation into proteins.
- Mechanisms: Key processes include alternative splicing, which allows for the production of different protein variants from a single gene; mRNA degradation that affects transcript stability; and silencing mechanisms mediated by microRNAs (miRNAs) and small interfering RNAs (siRNAs) that can inhibit translation.
- Translational Regulation:
- Definition: This regulation controls gene expression at the protein synthesis level, dictating the rate at which mRNA is translated into functional proteins.
- Mechanisms: It involves various factors that facilitate initiation and elongation during translation, ribosome binding to mRNA, and regulatory influences from miRNAs and siRNAs that can modulate translation efficiency.
- Post-translational Regulation:
- Definition: This process refers to the modifications that occur to proteins after they have been synthesized, impacting their functionality, localization within the cell, and overall stability.
- Mechanisms: Common modifications include phosphorylation, which can activate or deactivate protein functions; ubiquitination, leading to proteasomal degradation; and proteolytic processing, which can generate active protein forms from inactive precursors.
- Transcriptional Regulation:
Mechanisms of Eukaryotic Gene Regulation
Mechanisms of eukaryotic gene regulation are complex and multifaceted, enabling precise control over gene expression crucial for cellular function and organismal development. This regulation involves various processes that work in concert to fine-tune the expression of genes in response to internal and external cues.
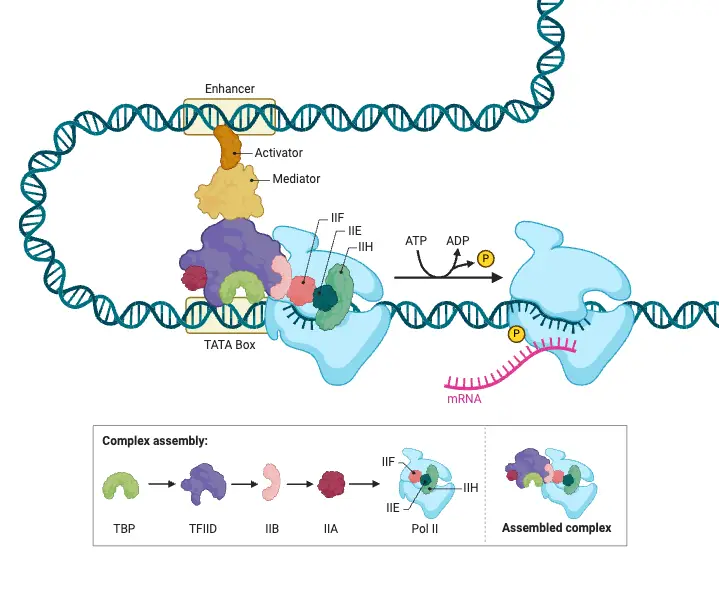
- Transcriptional Regulation:
- Mechanism: This involves the binding of transcription factors (TFs) to specific DNA sequences known as cis-regulatory elements located near the promoter region of genes.
- Types of Transcription Factors:
- Activators: These proteins enhance transcription by recruiting RNA polymerase II along with other coactivators to initiate gene expression.
- Repressors: Conversely, repressors inhibit transcription by either blocking the binding of activators or by recruiting corepressors, thus preventing the transcription machinery from accessing the gene.
- Chromatin Remodeling: This process modifies the chromatin structure to either expose transcription factor binding sites for activation or conceal them for repression, thereby influencing the accessibility of DNA for transcription.
- Post-transcriptional Regulation:
- Mechanisms:
- Alternative Splicing: This allows for the production of multiple mRNA transcripts from a single gene, thereby increasing protein diversity and functionality.
- mRNA Stability and Transport: The regulation of mRNA degradation rates and its subcellular localization are crucial for determining how much of a given protein is produced.
- MicroRNA (miRNA) and Small Interfering RNA (siRNA) Mediated Silencing: These small RNA molecules facilitate post-transcriptional gene silencing through mechanisms that either degrade mRNA or inhibit its translation.
- Mechanisms:
- Translational Regulation:
- Mechanisms:
- Initiation and Elongation Control: This involves the regulation of ribosome binding to mRNA and the subsequent movement of the ribosome along the mRNA strand during protein synthesis.
- miRNA and siRNA Influence on Translation: Similar to their role in post-transcriptional regulation, these small RNA molecules can target translation initiation, thus modulating protein production.
- Mechanisms:
- Post-translational Regulation:
- Mechanisms:
- Phosphorylation, Ubiquitination, and Other Covalent Modifications: These modifications can alter protein activity, localization, or stability, playing a critical role in controlling protein function after synthesis.
- Proteolytic Processing: This controlled degradation of proteins helps regulate their activity and can lead to the generation of active peptides necessary for various cellular processes.
- Mechanisms:
- Epigenetic Gene Regulation in Eukaryotes:
- Definition: This refers to heritable changes in gene expression that do not involve alterations in the underlying DNA sequence.
- Mechanisms:
- Epigenetic mechanisms control access to chromosomal regions through modifications that affect DNA packing around histone proteins.
- The addition or removal of chemical modifications to histones or DNA serves as signals for chromosomal regions to either open or close, thereby regulating gene accessibility.
- Coordination of Gene Expression in Eukaryotes:
- Complexity Due To:
- The absence of multigene operons necessitates the coordinated regulation of individual genes, requiring sophisticated regulatory networks.
- All nuclear DNA being wrapped in chromatin complicates access control, necessitating intricate mechanisms to facilitate or inhibit transcription.
- Chemical signals such as hormones, cytokines, and growth factors play vital roles in mediating timely changes in gene expression, allowing cells to respond appropriately to environmental stimuli.
- Complexity Due To:
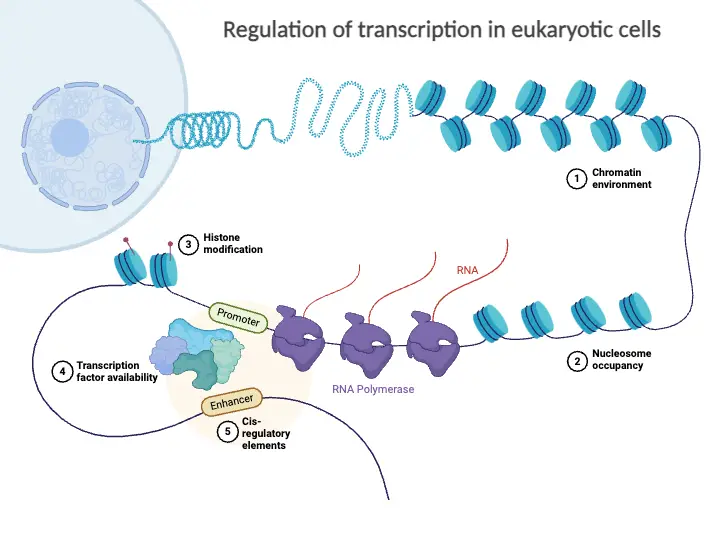
Regulatory Elements and Factors
Regulatory elements and factors are fundamental components in the intricate process of eukaryotic gene regulation. They play a crucial role in modulating gene expression, thereby influencing various cellular functions and responses to environmental changes. This discussion focuses on the definitions, types, and interactions of these essential elements and factors.
- Regulatory Elements:
- Definition: Regulatory elements are specific DNA sequences that serve as binding sites for regulatory proteins, influencing gene expression by facilitating or hindering the transcription process.
- Types:
- Promoters: These elements are located near the transcription start site of a gene and are essential for the recruitment of RNA polymerase II, the enzyme responsible for synthesizing mRNA.
- Enhancers: Enhancers can be positioned far from the promoter yet loop back to interact with the transcription initiation complex, significantly enhancing transcription levels.
- Silencers: Similar to enhancers, silencers are DNA sequences that can repress gene expression by inhibiting the transcription process.
- Insulators: These boundary elements prevent inappropriate activation of genes by blocking the interaction between enhancers and promoters, ensuring that gene expression is tightly controlled.
- Response Elements: Specific sequences that are recognized by transcription factors in response to external signals, such as hormone response elements, play a key role in mediating gene expression changes in reaction to environmental cues.
- Regulatory Factors:
- Definition: Regulatory factors are proteins that bind to regulatory elements to stimulate (activators) or inhibit (repressors) gene transcription.
- Types:
- Transcription Factors (TFs):
- General TFs: These are necessary for the transcription of most genes, such as TFIID and TFIIA, which are involved in the formation of the transcription initiation complex.
- Sequence-Specific TFs: These factors bind to specific DNA sequences to regulate particular genes, with notable examples including p53, a key player in DNA damage response, and NF-κB, involved in immune response.
- Coactivators and Corepressors:
- Coactivators: These proteins enhance the activity of transcription factors without directly binding to DNA, such as CBP/p300, which assist in forming the transcription complex.
- Corepressors: In contrast, corepressors inhibit transcription by interacting with transcription factors or modifying chromatin, examples include NCoR and SMRT.
- Transcription Factors (TFs):
- Interactions and Complex Formation:
- Transcription Factor-DNA Interactions: The sequence specificity of TFs is ensured by the unique shape and chemical properties of both the transcription factors and their respective binding sites on DNA.
- Protein-Protein Interactions: Interactions among transcription factors (TF-TF interactions) can either enhance or inhibit binding to DNA, affecting overall transcriptional outcomes. Additionally, TF interactions with coactivators and corepressors modify the transcriptional activity, facilitating a nuanced response to regulatory signals.
- Examples of Regulatory Elements and Factors in Action:
- The Lac Operon in Prokaryotes: While this example is from prokaryotes, it illustrates foundational principles of regulatory elements and factors that are relevant to understanding eukaryotic systems.
- The p53 Tumor Suppressor in Humans: This sequence-specific transcription factor responds to DNA damage, highlighting the significance of regulatory factors in disease prevention and cellular stress responses.
- The HIF-1 (Hypoxia-Inducible Factor 1): Under low oxygen levels, HIF-1α interacts with hypoxia response elements (HRE), orchestrating a transcriptional response that enables cells to adapt to hypoxic conditions.
Through the coordinated action of regulatory elements and factors, eukaryotic cells can precisely control gene expression, responding dynamically to a variety of internal and external stimuli. This regulatory architecture is crucial for maintaining cellular homeostasis and facilitating proper development and differentiation.
Techniques for Studying Gene Regulation
Studying gene regulation in eukaryotes involves a variety of techniques that allow researchers to explore the intricate mechanisms governing gene expression. Each method provides unique insights into the various layers of regulation, from molecular interactions to cellular responses. The following outlines essential techniques for investigating gene regulation, organized into distinct categories.
- Molecular Biology Techniques:
- DNA Footprinting and Gel Shift Assays:
- Purpose: These techniques identify protein-DNA interactions and transcription factor binding sites, crucial for understanding how gene expression is regulated at the transcriptional level.
- Methodology: DNA footprinting, such as DNase I footprinting, reveals protected regions of DNA where proteins bind, while gel shift assays (Electrophoretic Mobility Shift Assay, EMSA) demonstrate changes in DNA mobility upon protein binding.
- Chromatin Immunoprecipitation (ChIP) and ChIP-Seq:
- Purpose: ChIP techniques determine the binding of proteins, such as transcription factors, to specific genomic regions in living organisms.
- Methodology: This involves the immunoprecipitation of chromatin-associated proteins, followed by polymerase chain reaction (ChIP) or high-throughput sequencing (ChIP-Seq) to identify the specific DNA sequences associated with those proteins.
- DNA Footprinting and Gel Shift Assays:
- Biochemical and Biophysical Assays:
- Western Blotting and Co-Immunoprecipitation:
- Purpose: These methods are used to analyze protein expression levels and to investigate protein-protein interactions that play critical roles in gene regulation.
- Methodology: Western blotting detects specific proteins in a sample, while co-immunoprecipitation (co-IP) reveals interactions between proteins.
- Electrophoretic Mobility Shift Assays (EMSAs):
- Purpose: EMSAs are employed to study protein-DNA interactions, helping to characterize how proteins bind to regulatory regions of DNA.
- Methodology: This technique detects shifts in the electrophoretic mobility of DNA when it is bound by proteins, indicating binding events.
- Western Blotting and Co-Immunoprecipitation:
- Cell Biology and Imaging Techniques:
- Fluorescence Microscopy and Live Cell Imaging:
- Purpose: These techniques visualize the localization and dynamics of proteins within living cells, providing insights into gene regulation in real-time.
- Methodology: By tagging proteins with fluorescent markers, advanced microscopy techniques can track their movement and interactions.
- Flow Cytometry:
- Purpose: Flow cytometry quantifies protein expression levels in cell populations, allowing for the analysis of gene regulation effects across a large number of cells.
- Methodology: This technique measures fluorescence intensities from labeled cells as they pass through a laser, providing quantitative data on protein expression.
- Fluorescence Microscopy and Live Cell Imaging:
- Genomic and Computational Approaches:
- RNA Interference (RNAi) and CRISPR-Cas9 Gene Editing:
- Purpose: These approaches study the function of specific genes by knocking down or out gene expression, elucidating their roles in regulatory networks.
- Methodology: RNAi provides transient gene knockdown, while CRISPR-Cas9 facilitates permanent gene knockout, allowing researchers to assess the consequences of loss of function.
- Microarray Analysis and RNA-Seq:
- Purpose: These methods assess global gene expression changes, providing a comprehensive view of how gene regulation can vary under different conditions.
- Methodology: Microarrays utilize hybridization techniques, while RNA sequencing (RNA-Seq) employs high-throughput sequencing to measure gene expression levels across the entire transcriptome.
- RNA Interference (RNAi) and CRISPR-Cas9 Gene Editing:
- In Vivo Studies:
- Transgenic and Knockout Organisms:
- Purpose: These models are essential for examining gene function within the context of whole organisms, offering insights into the physiological relevance of specific genes.
- Methodology: Transgenic or knockout models can be generated using CRISPR-Cas9 or traditional genetic engineering methods, allowing for the exploration of gene regulation in living systems.
- Conditional Mutagenesis:
- Purpose: This technique enables the study of gene function in specific tissues or developmental stages, providing a nuanced understanding of gene regulation in diverse biological contexts.
- Methodology: Employing inducible or tissue-specific Cre recombinase systems allows researchers to manipulate gene expression spatially and temporally.
- Transgenic and Knockout Organisms:
The Consequences of Disrupted Gene Regulation in Eukaryotes
Dysregulation of gene regulation represents a significant area of concern in understanding various diseases. The delicate balance of gene expression is crucial for maintaining normal cellular functions. When this regulation is disrupted, it can lead to a multitude of health issues, including cancer, genetic disorders, and neurological conditions. The following points outline the causes of dysregulation and their associations with specific diseases.
- Causes of Dysregulation:
- Genetic Mutations:
- Point Mutations: These occur in regulatory elements or transcription factors, potentially altering their ability to bind DNA and control gene expression.
- Chromosomal Rearrangements: Such alterations can disrupt the regulatory landscapes of genes, leading to aberrant expression patterns.
- Epigenetic Alterations:
- DNA Methylation Changes: Aberrant methylation patterns can silence tumor suppressor genes or activate oncogenes, contributing to tumorigenesis.
- Histone Modifications: Changes in histone acetylation or methylation can modify chromatin structure, thereby influencing accessibility to transcription machinery and impacting gene expression.
- Environmental Factors:
- Toxins and Radiation Exposure: These external agents can induce genetic or epigenetic changes, disrupting normal regulatory processes.
- Infections: Pathogen-induced inflammatory responses can interfere with gene regulation, potentially leading to chronic dysregulation.
- Genetic Mutations:
- Diseases Associated with Dysregulation:
- Cancer:
- Tumor Suppressor Gene Silencing: For instance, the silencing of the p53 gene is commonly observed in various cancers, disrupting cell cycle regulation and apoptosis.
- Oncogene Activation: The HER2 gene is a well-known oncogene that, when overexpressed, contributes to breast cancer progression.
- Genetic Disorders:
- Down Syndrome (Trisomy 21): This condition results in the overexpression of genes located on chromosome 21, leading to developmental abnormalities.
- Fragile X Syndrome: This disorder is caused by the silencing of the FMR1 gene due to CGG repeat expansion, impacting cognitive functions.
- Neurological and Psychiatric Conditions:
- Alzheimer’s Disease: Dysregulation of genes involved in neuronal health, such as APP and PSEN1, plays a critical role in the pathology of Alzheimer’s disease.
- Schizophrenia: Epigenetic alterations can affect genes involved in brain development and function, contributing to the onset of schizophrenia.
- Immunological Diseases:
- Autoimmune Disorders: Conditions such as rheumatoid arthritis and lupus result from the dysregulation of immune response genes, leading to inappropriate immune reactions.
- Immunodeficiencies: Severe Combined Immunodeficiency (SCID) is caused by mutations in genes essential for the development and function of immune cells, resulting in heightened susceptibility to infections.
- Cancer:
- Therapeutic Interventions:
- Targeted Therapies:
- Small Molecule Inhibitors: These include kinase inhibitors used in cancer treatment, specifically targeting dysregulated signaling pathways.
- RNA Therapies: Techniques such as small interfering RNA (siRNA) and antisense oligonucleotides aim to correct or mitigate the effects of dysregulated gene expression.
- Gene Therapy:
- Gene Replacement or Editing: Methods like CRISPR-Cas9 can correct genetic defects by directly modifying the affected genes, providing potential cures for genetic disorders.
- Epigenetic Therapies:
- DNA Methyltransferase Inhibitors: Drugs such as azacitidine are used to treat myelodysplastic syndromes by reversing abnormal methylation patterns.
- Histone Deacetylase Inhibitors: Vorinostat, for example, is utilized in cutaneous T-cell lymphoma, acting to modify histone acetylation and restore normal gene expression.
- Targeted Therapies:
- Nature Structural and Molecular Biology 11, 812–815 (2004) doi:10.1038/nsmb820
- Struhl, K. Fundamentally different logic of gene regulation in eukaryotes and prokaryotes. Cell 98, 1–4 (1999)
- Sproul, D., Gilbert, N., & Bickmore, W. The role of chromatin structure in regulating the expression of clustered genes. Nature Reviews Genetics 6, 775–781 (2005) doi:10.1038/nrg1688
- Alberts, B. et al. (2002). Molecular Biology of the Cell. 5th edition. Garland Science.
- Green, M. R., & Sambrook, J. (2012). Molecular Cloning: A Laboratory Manual. 4th edition. Cold Spring Harbor Laboratory Press.
- Wingren, C., & Borrebaeck, C. A. K. (2009). High-throughput proteomics for biomarker discovery. Expert Review of Proteomics, 6(1), 43-54. doi: 10.1586/epr.08.67
- Strachan, T., & Read, A. P. (2018). Human Molecular Genetics. 5th edition. Garland Science.
- Alberts, B. et al. (2002). Molecular Biology of the Cell. 5th edition. Garland Science.
- Esteller, M. (2008). Epigenetics in Cancer. New England Journal of Medicine, 358(11), 1148-1159. doi: 10.1056/NEJMra072067
- https://www.etsu.edu/uschool/faculty/tadlockd/documents/18_detaillectout.pdf
- https://www.csun.edu/~cmalone/pdf360/Ch20-1%20euk%20gene%20reg.pdf
- https://bio.libretexts.org/Courses/University_of_Arkansas_Little_Rock/Genetics_BIOL3300_(Leacock)/Genetics_Textbook/06%3A_Regulation_of_Gene_Expression/6.02%3A_Eukaryotic_Gene_Regulation