What are Enhancers and silencers?
- Enhancers and silencers are key regulatory elements within genomes that control gene expression in metazoans. These elements play critical roles in ensuring that genes are turned on or off at the right times and in the right cells, which is essential for proper development and cellular function.
- Enhancers are short DNA sequences containing binding sites for transcription factors. These sequences can activate their target genes regardless of their position relative to the gene they regulate, even acting over long distances within the same chromosome (in cis) or across different chromosomes (in trans). Enhancers function by recruiting transcription factors and other proteins that facilitate the assembly of the transcriptional machinery at the gene’s promoter, thereby boosting gene expression. They are essential in driving high levels of gene expression during processes like development and differentiation.
- On the other hand, silencers serve the opposite role—they suppress gene expression. Silencers can either inhibit transcription directly or restrict it to specific regions of chromatin, acting as insulators. By blocking the access of transcription factors or preventing the formation of transcription complexes at a gene’s promoter, silencers ensure that certain genes remain inactive when they are not needed. This regulation is particularly important in maintaining cell identity and preventing the inappropriate activation of genes.
- The relationship between enhancers, silencers, and their target promoters is finely tuned and influenced by the three-dimensional structure of chromatin. Genes are not randomly organized within the nucleus but are arranged in loops, rosettes, and other higher-order structures that bring regulatory elements into contact with their target genes. Enhancers and silencers work within this complex 3D environment, ensuring that genes are expressed in the right place and at the right time.
- Interestingly, despite their opposing effects on gene expression, enhancers and silencers may share some functional similarities. Both elements likely work by interacting with specific regions in the nucleus known as transcription factories, where active gene transcription occurs. These interactions may involve the production of non-coding RNA (ncRNA), which could play a role in regulating the activity of both enhancers and silencers.
What is Enhancer?
Enhancers play a critical role in regulating gene expression in both prokaryotes and eukaryotes. They are specific regions of DNA that, despite their distance from the genes they influence, serve as crucial elements for the transcriptional activation of genes. Below is a detailed breakdown of what enhancers are, how they function, and their significance in genetics.
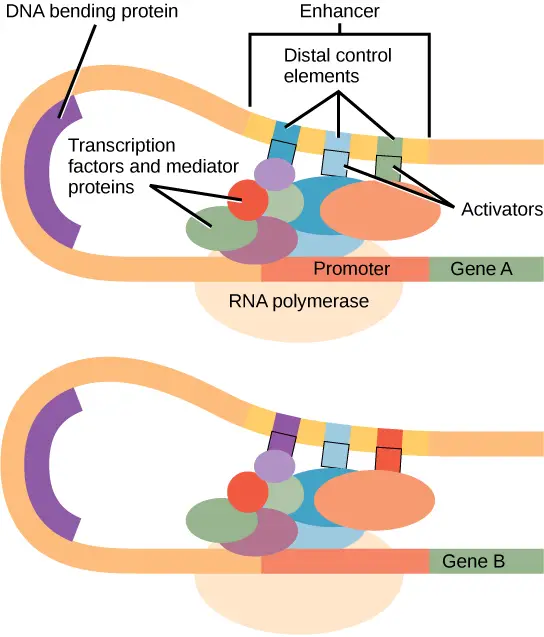
- Definition and General Features:
- Enhancers are short DNA sequences, typically ranging from 50 to 1,500 base pairs (bp), that can significantly increase the probability of gene transcription.
- These regions act as binding sites for proteins, known as transcription factors or activators, which are necessary for initiating gene expression.
- Unlike promoters, which are usually situated near the gene, enhancers can be located far from the gene they regulate, sometimes up to 1 million base pairs away, and may be found either upstream or downstream of the gene.
- In the human genome alone, there are hundreds of thousands of these enhancers.
- Cis-acting Nature:
- Enhancers are classified as “cis-acting” elements, meaning they influence the transcription of genes located on the same chromosome.
- They operate by facilitating the assembly of the transcription machinery at the gene’s promoter through protein-DNA interactions.
- Role in Chromatin Structure:
- Enhancers are embedded within the complex structure of chromatin, often located in regions of “open” chromatin that are accessible to transcription factors. This accessibility is indicated by sensitivity to enzymes like DNase I.
- Specific chemical markers on histone proteins (such as H3K4me1, H3K4me2, and H3K27ac) flank these regions, aiding in their identification and activity.
- Functional Variability and Flexibility:
- Enhancers exhibit considerable flexibility. They can function regardless of their orientation and can activate genes either in cis (within the same chromosome) or trans (across different chromosomes).
- They can be located in diverse regions, such as gene deserts, introns, or untranslated regions.
- Evolutionary Perspective:
- Although sequence conservation across species can sometimes predict enhancer function, there are cases where enhancers governing similar gene expression patterns in different species show little to no sequence similarity.
- Identification of Enhancers:
- In addition to chromatin accessibility, enhancers are often marked by the binding of proteins like p300 (a histone acetyltransferase), and RNA polymerase II (RNAPII), further assisting in their detection.
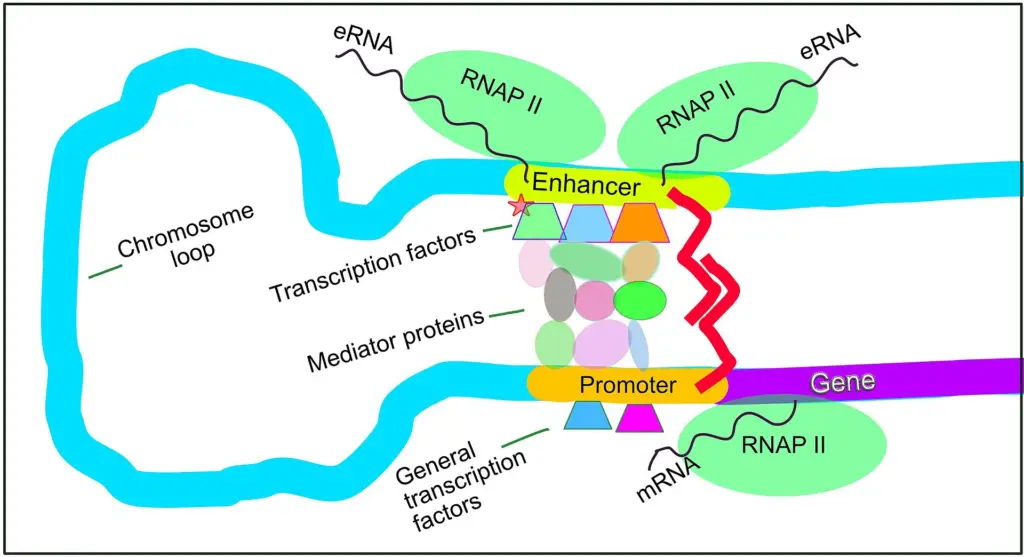
- Specific modifications of histones around enhancer regions and interactions with other chromatin-bound proteins like Mediator subunits and CTCF are common markers.
- Transcription of Enhancers:
- Enhancers are transcribed into non-coding RNA molecules, known as enhancer RNAs (eRNAs). These eRNAs do not encode proteins but seem to play a role in stabilizing the interaction between enhancers and promoters, facilitating transcription.
- The production of eRNAs has been observed in a number of gene loci, including the β-globin locus and the Arc promoter region.
- Example of Enhancer Action:
- One of the most well-studied examples of enhancer function is seen in the β-globin locus, where an enhancer region, termed the locus control region (LCR), is located far upstream from the promoter it regulates. This enhancer interacts with its target promoter through chromatin loops, allowing transcription to proceed.
- Enhancers and ncRNA (Non-Coding RNA):
- Beyond eRNAs, another class of non-coding RNA, known as long intergenic non-coding RNA (lincRNA), has been associated with enhancer function. These lincRNAs, typically longer than 200 nucleotides, have been shown to influence the transcription of nearby genes.
- For instance, HOTTIP, a lincRNA from the HOXA locus, facilitates the activation of multiple HOXA genes by promoting chromatin looping.
- Enhancers in Medical Research and Biotechnology:
- Recent studies have linked enhancers to medical conditions, such as myelosuppression. Furthermore, advances in artificial intelligence have led to the design of synthetic enhancers, which have been applied in both in vitro (cell line) and in vivo (animal) systems since 2022.
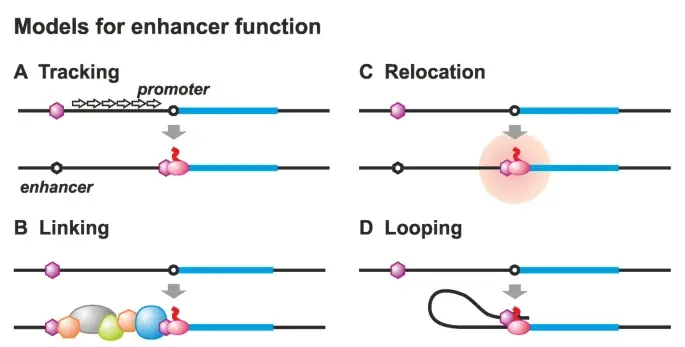
Location of Enhancer
nhancers are crucial regulatory elements in eukaryotic cells that significantly influence gene expression. Their locations within the genome are diverse, and their spatial relationships to target genes enable them to perform their roles effectively. Below are key points that describe the various locations and characteristics of enhancers:
- Proximity to Target Genes:
- Enhancers may reside both upstream and downstream of the genes they regulate. This flexibility in positioning allows for significant regulatory control over transcription.
- Importantly, enhancers do not need to be located near the transcription initiation site to exert their effects. Some enhancers have been identified hundreds of thousands of base pairs away from the start site of transcription, demonstrating their ability to influence distant genes.
- Interaction with Promoters:
- Although enhancers are not situated directly on the promoter region of a gene, they play an essential role by interacting with general transcription factors and RNA polymerase II.
- This interaction occurs through the folding of the chromatin, which spatially brings the enhancer into close contact with the promoter, facilitating the recruitment of transcription machinery.
- Location Within Introns:
- Enhancers can be found within the intronic regions of genes. This positioning allows them to influence the transcription of the associated gene without being part of the coding sequence.
- The presence of enhancers in introns can also explain why polymorphisms in these regions may affect gene expression despite not being translated into proteins.
- Orientation and Flexibility:
- An enhancer’s orientation is not fixed; it can function effectively even if its orientation is reversed. This property indicates a level of robustness in enhancer functionality.
- Additionally, enhancers can be excised from their original locations and inserted elsewhere in the genome without losing their ability to regulate gene transcription.
- Enhancers Across Different Genes:
- Enhancers may also be located in the exonic regions of unrelated genes, allowing for a complex network of regulatory interactions across the genome. This aspect emphasizes the interconnected nature of gene regulation in eukaryotic cells.
- Moreover, some enhancers are capable of acting on genes situated on different chromosomes, which further illustrates the expansive regulatory capabilities of these elements.
- Identification through Coactivator Binding:
- Enhancers are often bound by coactivators such as p300 and CBP (CREB-binding protein). These interactions can be studied through techniques like ChIP-seq, which allows researchers to predict the locations of enhancers based on coactivator binding patterns.
Examples of Enhancer
Below are notable examples of enhancers, illustrating their critical roles in genetic regulation.
- HACNS1:
- HACNS1, also referred to as CENTG2, is an enhancer located in the Human Accelerated Region 2 of the genome. It is believed to have played a significant role in the evolution of human-specific traits, particularly those related to the structure and function of the hand and foot.
- This enhancer is thought to have contributed to the development of the uniquely opposable human thumb, a key feature that distinguishes humans from other primates. Furthermore, HACNS1 may have influenced modifications in the ankle or foot, enabling bipedal locomotion—an essential aspect of human evolution.
- Among the 110,000 gene enhancer sequences identified in the human genome, HACNS1 shows the most pronounced evolutionary changes following the divergence of humans from their common ancestors with chimpanzees. This highlights its potential role in driving key evolutionary adaptations in humans.
- GADD45G Enhancer:
- Another interesting example is an enhancer located near the gene GADD45G, which plays a role in regulating brain growth in various mammals, including chimpanzees and mice. However, in humans, this enhancer appears to be inactive or absent, contributing to notable differences in brain development across species.
- In chimpanzees and mice, the GADD45G enhancer is active in areas of the brain responsible for forming the cortex, ventral forebrain, and thalamus. Its activation in these regions may suppress further neurogenesis, limiting brain growth. The absence or reduced activity of this enhancer in humans is thought to have allowed for the expansion of certain neuronal populations, contributing to the increased size and complexity of the human forebrain.
- The loss of this enhancer in humans could explain the differences in brain development between humans and other mammals, emphasizing the role of enhancers in shaping species-specific traits, particularly in relation to neurological evolution.
What is silencer?
Silencers are specialized DNA sequences that play a crucial role in gene regulation by inhibiting transcription. Unlike enhancers, which promote gene expression, silencers function to repress genes, preventing them from being expressed as proteins. This repression is essential for various biological processes, including cellular differentiation and the control of the cell cycle.
- Function and Mechanism:
- Silencers are DNA sequences that bind to regulatory proteins known as repressors. When a repressor protein attaches to the silencer region of DNA, it blocks RNA polymerase from initiating transcription. This prevents the DNA sequence from being transcribed into RNA, which is a necessary step for protein production.
- The primary function of silencers is to prevent genes from being expressed at times when their activity is not required. By controlling when specific genes are turned off, silencers play an important role in maintaining proper cellular function and development.
- Role of Noncoding RNAs in Silencing:
- Accumulating evidence shows that both long and short RNA molecules can be involved in silencing gene expression. For example, antigene RNAs (agRNAs) are small RNA molecules that target promoters and regions downstream of genes to inhibit transcription.
- Additionally, microRNAs (miRNAs), which are typically 20 to 22 nucleotides in length, regulate gene expression at the post-transcriptional level but can also impact transcriptional initiation. These miRNAs may localize in the nucleus and interact with repressors to further enhance the gene silencing process.
- Involvement of Polycomb Complexes:
- Silencers often recruit specific protein complexes, such as Polycomb Repressive Complexes (PRC1 and PRC2), which play key roles in maintaining transcriptional repression. For instance, PRC2 catalyzes the trimethylation of histone H3 at lysine 27 (H3K27me3), a modification that helps recruit PRC1 and reinforces chromatin compaction, thereby preventing the transcription of target genes.
- The interaction of these Polycomb complexes with noncoding RNAs like HOTAIR contributes to gene silencing. For example, overexpression of HOTAIR in breast cancer cells results in the recruitment of PRC2 to many genetic loci, effectively silencing more than 850 target genes.
- Three-Dimensional Chromatin Structure:
- The spatial arrangement of chromatin is crucial for the functioning of silencers. In many cases, the three-dimensional structure of the genome brings silencers into close proximity with the genes they regulate. This interaction enables silencers to compact chromatin or rearrange it in a way that makes the gene inaccessible to transcription factors, ensuring that transcription is effectively blocked.
- In processes like X chromosome inactivation, the ncRNA Xist binds to PRC2, leading to the propagation of silencing marks across the inactive X chromosome. This involves large-scale chromatin compaction and prevents the transcription of genes along the silenced chromosome.
- Regulation and Plasticity:
- Gene silencing is not always permanent. Silencers can be deactivated, allowing the gene to return to an active state. This process involves the eviction of silencing proteins like Polycomb complexes, restoring accessibility to the gene and allowing transcription to resume.
- This dynamic regulation is critical during development and in response to environmental signals. The ability to toggle between active and silenced states ensures that gene expression is tightly controlled based on the needs of the cell.
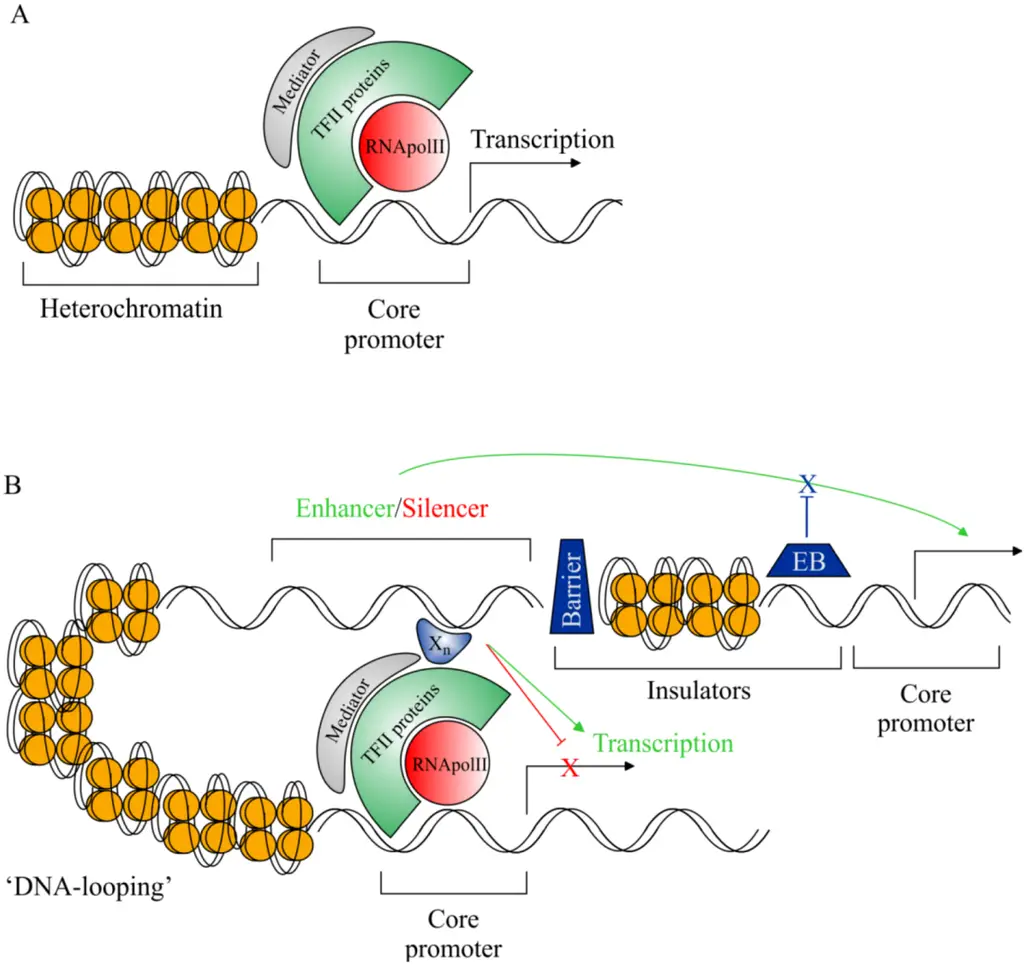
Locations of Silencers within the Genome
Silencers are crucial regulatory elements in the genome that function to repress the transcription of specific genes. Their locations within the DNA are varied, allowing for significant regulatory control over gene expression. The following points outline the diverse locations of silencers and their implications for transcription regulation:
- Common Upstream Position:
- The most frequent location of silencer elements is upstream of the target gene. This positioning allows silencers to effectively interact with transcription machinery and repress transcription.
- The distance of a silencer from the gene can vary significantly, typically ranging from approximately 20 base pairs (bp) to 2000 bp upstream of the transcription start site. This variability provides flexibility in regulatory interactions.
- Downstream Locations:
- Besides being found upstream, silencers can also be located downstream of the gene they regulate. This placement further contributes to the intricate control of transcription.
- Some silencers may exist within the introns of a gene, thereby influencing transcription while being part of the non-coding regions of the gene.
- Exonic and Intronic Presence:
- Silencers can also be found within the exonic regions of a gene, allowing them to directly participate in the regulation of the coding sequence.
- This presence within introns and exons highlights the multifunctional nature of these elements, enabling them to exert control over transcription from various positions.
- 3′ Untranslated Region:
- Silencers have also been identified within the 3′ untranslated region (3′ UTR) of mRNA. This region, while not translated into protein, plays a significant role in the post-transcriptional regulation of gene expression.
- The inclusion of silencers in the 3′ UTR suggests a mechanism by which gene expression can be modulated after transcription has occurred.
Examples of silencer
Below are key examples of silencers, illustrating their function in gene repression.
- Polycomb Repressive Complex (PRC) Silencers:
- Polycomb repressive complexes (PRC1 and PRC2) are well-known for their involvement in gene silencing through interaction with specific noncoding RNA transcripts. These complexes target silencing elements to regulate gene repression in a range of biological contexts.
- In CD4+ T-cells and embryonic stem (ES) cells, for instance, PRC2 catalyzes the trimethylation of histone H3 at lysine 27 (H3K27me3), which recruits PRC1. This recruitment prevents chromatin remodeling at the targeted loci, ensuring that these regions remain transcriptionally inactive.
- A notable example of this mechanism is the interaction between PRC2 and HOTAIR, a long noncoding RNA. In this case, HOTAIR drives the repression of genes within the HOXC locus by recruiting PRC2 and promoting H3K27 trimethylation, leading to chromatin compaction and gene silencing.
- X-Chromosome Inactivation:
- One of the most well-studied examples of silencing is X-chromosome inactivation (XCI) in females, a process in which one of the two X chromosomes is transcriptionally silenced to ensure dosage compensation. The noncoding RNA Xist plays a central role in this process.
- Xist binds to the X chromosome that will be inactivated, recruiting PRC2 to establish H3K27 trimethylation. This leads to chromatin compaction and the silencing of gene expression across the entire chromosome.
- The three-dimensional conformation of the chromatin is critical for efficient silencing, as it results in the propagation of PRC1 binding and the formation of a repressive chromatin structure that remains inactive throughout the cell’s life.
- miRNA-Associated Silencers:
- MicroRNAs (miRNAs) are small RNA molecules that not only regulate gene expression post-transcriptionally but can also act as silencers at the transcriptional level. Mature miRNAs, which are usually found in the cytoplasm, have also been identified in the nucleus, where they modulate transcriptional initiation.
- For example, miRNAs that target the progesterone gene promoter have been shown to decrease RNA polymerase II occupancy, thereby reducing transcription. This silencing is associated with increased levels of H3K9 dimethylation (H3K9me2), a repressive histone modification, in an Argonaute 2 (Ago-2)-dependent manner.
- These findings suggest that miRNAs can function as silencers by promoting repressive chromatin states, thereby inhibiting the transcription of their target genes.
- Antigene RNAs (agRNAs):
- Antigene RNAs (agRNAs) are another class of small RNA molecules that can silence gene expression by targeting specific promoter regions. By binding to these regions, agRNAs inhibit the transcription of genes such as those encoding progesterone, the androgen receptor, and cyclooxygenase-2.
- The repression induced by agRNAs is associated with changes in chromatin structure, including DNA methylation at the promoter and modifications of histones that favor a repressed state. These small RNA molecules provide a mechanism for precise control of gene expression at the transcriptional level.
Functions of Enhancers and silencers
Enhancers and silencers are pivotal regulatory elements in gene expression, functioning as key players in the intricate control of transcription. Each element serves distinct but complementary roles in the regulation of gene activity. Below is a detailed exploration of their respective functions:
- Functions of Enhancers:
- Activation of Transcription:
- Enhancers facilitate the binding of transcription factors and coactivators to promote the transcription of target genes. They can significantly increase the likelihood of transcription initiation by RNA polymerase II.
- Spatial Flexibility:
- Enhancers can be located far from the genes they regulate, sometimes hundreds of thousands of base pairs away. Despite this distance, they achieve their effects by looping the DNA, bringing them into proximity with the promoter region.
- Binding of Specific Proteins:
- Enhancers are bound by specific transcription factors, including activator proteins, which interact with the mediator complex. This interaction is crucial for recruiting RNA polymerase II and the general transcription factors necessary for initiating transcription.
- Regulation Across Chromosomes:
- Enhancers have the unique ability to influence genes located on different chromosomes, highlighting their versatility in gene regulation. This trans-regulatory capability allows for complex regulatory networks within the genome.
- Orientation Independence:
- The functionality of enhancers is maintained even when their orientation is reversed or when they are excised and relocated within the genome. This characteristic underlines the robustness of enhancer action in regulating gene expression.
- Activation of Transcription:
- Functions of Silencers:
- Repression of Transcription:
- Silencers serve to inhibit the transcription of target genes by binding to repressor proteins. This binding prevents the assembly of the transcription machinery at the promoter region, effectively silencing gene expression.
- Specificity in Regulation:
- Silencers provide specificity in gene regulation by selectively targeting certain genes for repression, allowing cells to control the expression of specific proteins in response to developmental signals or environmental changes.
- Varied Locations:
- Silencers can be found upstream, downstream, or even within introns and exons of the genes they regulate. Their diverse locations allow for nuanced control over transcription, depending on the context of gene expression.
- Interaction with Transcription Factors:
- Like enhancers, silencers also interact with transcription factors, but in this case, they bind repressor proteins that antagonize the function of activators. This antagonistic relationship is vital for maintaining gene expression patterns.
- Role in Fine-Tuning Gene Expression:
- Silencers are crucial for the fine-tuning of gene expression, ensuring that genes are expressed at appropriate levels in response to various cellular conditions. This regulation is important for processes such as development, differentiation, and cellular responses to stimuli.
- Repression of Transcription:
- Kolovos, P., Knoch, T.A., Grosveld, F.G. et al. Enhancers and silencers: an integrated and simple model for their function. Epigenetics & Chromatin 5, 1 (2012). https://doi.org/10.1186/1756-8935-5-1
- Eukaryotic Gene Regulation – Transcriptional Enhancers and Repressors. (2023, November 1). https://bio.libretexts.org/@go/page/13361
- Ogbourne S, Antalis TM. Transcriptional control and the role of silencers in transcriptional regulation in eukaryotes. Biochem J. 1998 Apr 1;331 ( Pt 1)(Pt 1):1-14. doi: 10.1042/bj3310001. PMID: 9512455; PMCID: PMC1219314.
- https://en.wikipedia.org/wiki/Enhancer_(genetics)
- https://www.ndsu.edu/pubweb/~mcclean/plsc431/geneexpress/eukaryex4.htm
- https://en.wikipedia.org/wiki/Silencer_(genetics)