Differential centrifugation (or the differential velocity method) is one of the most prevalent techniques applied in biochemistry and cell biology regarding the separation of organelles and other subcellular particles according to their sedimentation rates. It is available not only for separate microscopic analyses, but also as a useful technique for preliminary remediation of suspended nonliving particles such as nanoparticles, colloidal particles or even viruses. As an example, tissue specimens that are subjected to differential centrifugation in order to study cell biological phenomena such as organelle behavior are first subjected to a de-lysing process in which the cell membranes are permeablized so that organelles and cytosol are released. This lysate is subsequently centrifuged multiple times. Particles that are solvable with a given centrifugal force for a specific time frame end up at the lower section of the centrifugal tube forming an elongated type of mosec of muscle structure called “pellet”.
After every centrifugation cycle, the supernatant is collected, often referred to as the non-pelleted solution. It is reseparated with increased centrifugal force or for a longer duration. The sedimentation rate is useful, however, it only allows for the separation of crude particles. Differential centrifugation can easily aid in this process. Further separations, more refined than that, can be accomplished by equilibrium centrifugation with the use of density gradients. This means that differential centrifugation is a process which is repeatedly performed with stronger and stronger centrifuge forces to collect previously supernatented particles. Organelles from differential centrifugation can remain highly functional if they are not subjected to isolation denaturing conditions.
Principle of Differential centrifugation
Differential centrifugation uses the rate of sedimentation as a function of the particle’s size and density. The application of greater centrifugal forces achieves sedimentation of larger molecules first. Various particles continue to sediment based on the parameters of each centrifugation cycle including the length of time the centrifuge is run as well as the size and density of each individual particle. The largest collection of particles remains intact within the tube, instead of decanting out with place holding structures that are lesser in size above it. Hence, larger molecules sediment faster and require less centrifugal force while smaller molecules require more time and force to achieve sedimentation. Particles that have lower density than the medium tend to float rather than sink.
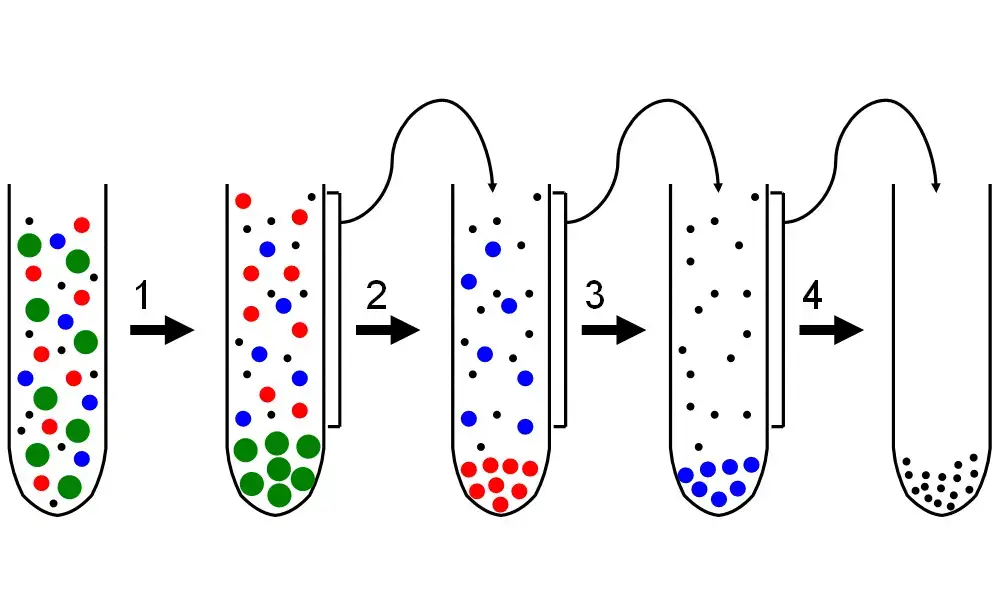
Differential centrifugation Protocol
The organelles of sub-cellular origin such as the nucleus, mitochondria, lysosomes, and microsomes can be isolated from a liver homogenate using differential centrifugation. The procedure consists of these steps:
- A 10% liver solution in 0.25 milliliters of sucrose is prepared.
- Centrifugation is done at 1000g for 10min.
- The pellet is separated from the sedimented nucleus.
- The supernatant from the previous step is decanted and centrifuged at 3300g for 10min.
- Separation of sedimented pellet containing mitochondria.
- The supernatant from previous step is centrifuged at 16300g for 20min.
- Separation of pellet that has been sedimented and contains lysosomes.
- The supernatant that was decanted in the previous step is centrifuged at 105000g for 60 minutes.
- The pellet is separated from the sediment containing microsomes.
- The supernatant collected from this stage is cell-free cytosol.
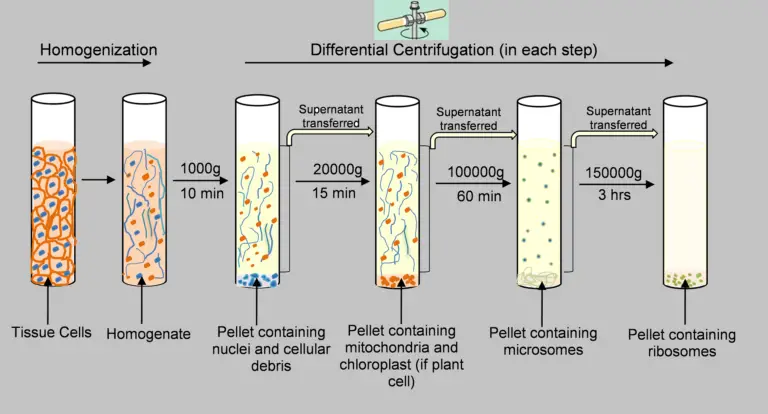
The isolation of organelles in sub-cellular form is a fundamental procedure in numerous biochemical research labs through this use of different centrifugation methods. A diagrammatic representation of steps-wise separation of sub-cellular organelles using an homogenate from the liver is provided in the following figure.
Fractionation by differential centrifugation
When preparing a cell homogenate, tissue, or whole cells together with their nuclei and large debris will accumulate in the unbroken pellet (U1) after centrifugation at a low speed (400-500 x g) for 10 minutes. The low speed pellet is often referred to as ‘nuclear pellet’. It is obtained after spinning at a moderately rapid speed for 10 minutes when forces between 10,000 x g and 220,000 x g are exerted. This is potent enough to destroy both lysosomes and peroxisomes but sufficient for the destruction of mitochondria. Hence, the second part of the traditional cell fractionation is known as the ‘mitochondrial pellet’.
Differential centrifugation requires the use of an ultracentrifuge. An ultracentrifuge is built to rotate rotors at very high speeds in order to produce tremendous gravitational forces. To prevent excessive heat from the friction with the air, the chamber has to be vacuumed. Indeed, many rotors designed to be used in an ultracentrifuge spin in the vacuum, Therefore, they were not designed aerodynamically. When spinning an ultracentrifuge at a force of 80,000 G x for one hour, microsomal pellets are formed. Microsomes are membrane-bound vesicles which contain fragments of the cell membrane and the endoplasmic reticulum. When membrane fragments are suspended in water and break apart, they form vesicles and upon closer inspection one can see a multitude of vesicles of various sizes. Based on the varying amounts of protein within the vesicles it is possible to distinguish them on the basis of buoyant density. But this will be the subject of a different paper.
You may centrifuge at 150,000xg for several hours and you should obtain ribosomes, along with what is probably the largest macromolecular structure. The leftover supernatant consists of the soluble fractions of the cells like salts, small macromolecules, precursor compounds and dissolved gases.
Sample preparation for Differential centrifugation
- Tissue or cells should be collected and immediately put on ice.
- The sample will be washed with cold saline followed by removal of moisture.
- Prepare a blend of isotonic salts, tris – HCl and sucrose, which will be used for tissue homogenization. Add protease inhibitors right before use.
- Place the sample in a cold homogenizer, then add buffer and disrupt the cells via a dounce homogenizer or a motorized pestle.
- Filter out the homogenate through several layers of cheescloth to remove large particulates and debris.
- Centrifuge the homogenate at 600-800 x g for 5-10 minutes. The supernatant will contain most organelles and the pellet will contain unbroken cells and nuclei.
- If preferred, wash the nuclear pellet with additional buffer and resuspend for further analysis.
- Transfer the supernatant into a new tube and spin it at roughly 10,000 x g for 15-30 minutes. Mitochondria lysosomes and peroxisomes will be pelleted.
- If necessary wash and resuspend the mitochondrial pellet in an appropriate buffer.
- Transfer the remaining supernatant into another tube and centrifuge at 100,000 x g for 1-2 hours with an ultracentrifuge. The pellet will be contianing small vesicles referred to as microsomes and the supernatant the cytosolic fraction.
- To increase purity, wash the pellets with buffer and re-centrifuge.
- All fractions should either be frozen immediately or stored long-term at –80 °C.
What is Equilibrium (isopycnic) sedimentation?
Equilibrium (isopycnic) sedimentation is a technique of centrifugation that separates sample components exclusively on the basis of their size. This method involves the use of a pre-prepared gradient with the sample applied on top of it, which is usually prepared with sucrose. During centrifugation, every particle in the sample is displaced throughout the gradient until their density exactly corresponds to the density of the surrounding medium. At the ‘isopycnic point’, the particle becomes motionless because it is no longer acted upon by any net force. This method is particularly helpful in the purification of complicated heterogenous mixtures such as the separation of different organelles or macromolecules because only the density differences of the particles are mechanized with no regard to other characteristics like the size or the shape of the particles.
Differential centrifugation vs density gradient centrifugation
| Aspect | Differential Centrifugation | Density Gradient Centrifugation |
|---|---|---|
| Principle | Sequential spins at increasing speeds to pellet components based on size and sedimentation rate. | Particles migrate in a pre-formed density gradient until they reach a position where the medium’s density equals their own (isopycnic point). |
| Separation Basis | Size and density differences combined with sedimentation speed. | Purely based on buoyant density. |
| Procedure Complexity | Relatively simple and fast; does not require gradient preparation. | More complex; requires the creation of a continuous or discontinuous density gradient. |
| Purity of Fractions | Yields crude fractions that may contain contaminants; often requires further purification. | Produces highly purified fractions as particles settle at their exact density. |
| Typical Applications | Initial cell fractionation (e.g., separating nuclei, mitochondria, and microsomes). | Fine separation of organelles, viruses, or macromolecules with similar sedimentation rates. |
What is the difference between centrifugation and differential centrifugation?
| Aspect | Centrifugation | Differential Centrifugation |
|---|---|---|
| Definition | A general technique that uses centrifugal force to separate particles based on size, shape, and density. | A specific type of centrifugation that involves sequential spins at increasing speeds to fractionate cell components based on their sedimentation rates. |
| Process | Typically a single or simple spin that separates a mixture into a pellet and supernatant. | Involves multiple steps with progressively higher speeds, yielding several pellets enriched with different cellular organelles. |
| Separation Basis | Separation is based on physical properties such as mass, density, and size. | Separation exploits differences in sedimentation rates among cellular components under varying centrifugal forces. |
| Outcome | Results in a basic division (pellet vs. supernatant). | Produces distinct fractions, each enriched in specific organelles or subcellular structures. |
| Applications | Used for general clarification, sedimentation of cell debris, or simple particle separation. | Commonly applied in cell biology for isolating nuclei, mitochondria, lysosomes, and other organelles from homogenized cells. |
Application of Differential centrifugation
- Removing organelles such as nuceli, mitochondria, lysosomes, and peroxisomes from homogenized cells for further studies.
- Recovery of the cytosol and microsomal (membrane) fractions for biochemical and proteomic studies.
- Prepartion of electron microspope samples by filtration for enrichment of particular subcellular components.
- Isolation of viruses or extracellular vesicles from biological mixtures of higher complexity.
- Act as a first stage in cell fractionation process before the use of more sophisticated separation techniques like density gradient centrifugation.
Advantages of Differential centrifugation
- It is a simple and rapid technique that does not require complex equipment or procedures.
- It allows for the initial separation of cellular components based on differences in size and density.
- It is cost-effective and scalable, making it suitable for processing large sample volumes.
- It produces crude fractions that can serve as a starting point for further, more refined purification methods.
- It does not require the preparation of a density gradient, which simplifies the overall process.
Limitations of Differential centrifugation
- Produces crude fractions that may contain significant contamination due to overlapping sedimentation rates.
- Cannot fully separate organelles or particles with similar sizes and densities.
- Multiple centrifugation steps are required, which can be time-consuming and labor-intensive.
- High-speed spins may damage delicate cellular structures or cause aggregation.
- Variability in sample preparation can lead to inconsistent results.
How to increase orgenlle purity after differential centrifugation?
- After differential centrifugation, perform an additional purification step by layering the crude fraction on a density gradient (using sucrose, Percoll, or iodixanol) and centrifuge. This allows the organelles to migrate to their equilibrium (isopycnic) position based solely on buoyant density.
- Wash the pelleted organelles by resuspending them in a fresh buffer and re-centrifuging to remove contaminants and loosely associated debris.
- Optimize centrifugation parameters (speed, time, and temperature) during both the differential and gradient steps to minimize aggregation or damage.
- If necessary, use immunoaffinity methods with antibodies specific to organelle markers to further purify the target organelle from any remaining contaminants.
FAQ
What does the process of differential centrifugation achieve?
The process of differential centrifugation separates different components of a biological sample based on their size, density, and shape. It involves subjecting the sample to high levels of centrifugal force, which causes larger, denser components to sediment to the bottom of the tube, while smaller, less dense components remain suspended in the supernatant. The process of differential centrifugation can be used to isolate specific cell types, organelles, proteins, or other subcellular structures from a complex mixture.
Differential centrifugation is a useful technique for purifying and isolating specific components of a biological sample for further study or analysis. It can be used to prepare samples for a wide range of applications, including microscopy, biochemistry, molecular biology, and cell biology. Differential centrifugation is a relatively simple and cost-effective method that is widely used in research and other scientific applications.
What is left in the supernatant of differential centrifugation?
During the process of differential centrifugation, larger, denser components of a biological sample sediment to the bottom of the tube, while smaller, less dense components remain suspended in the supernatant. The specific components that remain in the supernatant will depend on the specific sample being processed and the conditions used for the centrifugation.
In general, the supernatant of differential centrifugation may contain a variety of components, including cells, organelles, proteins, and other small molecules. The components that remain in the supernatant may be further separated and purified using additional techniques, such as density gradient centrifugation, chromatography, or immunoprecipitation.
Overall, the supernatant of differential centrifugation contains a mixture of smaller, less dense components that have not sedimented during the centrifugation process. These components may be of interest for further study or analysis, and can be further purified using additional techniques as needed.