What is Chromosome Mapping?
Chromosome mapping is a pivotal technique in genetics that determines the relative positions of genes on a chromosome. This process involves creating maps that systematically organize and elucidate genetic information on chromosomes. These maps not only depict the positions of genes but also the distances between them, scaled to a specific unit. The primary objective of chromosome mapping is to comprehend the organization, structure, and function of the genome.
Thomas Hunt Morgan, a renowned geneticist, played a crucial role in shaping the contemporary understanding of chromosome mapping. His groundbreaking experiments with fruit flies (Drosophila) unveiled evidence for genetic linkage. One of Morgan’s significant findings was the location of the white eye gene on the X chromosome of Drosophila. This discovery paved the way for the identification and mapping of other X-linked genes, revealing a linear arrangement of genes on the chromosome.
Alfred H. Sturtevant, while working as an undergraduate in Morgan’s laboratory, pioneered the method for mapping chromosomes in 1911. He utilized data from Drosophila crosses and harnessed the phenomenon of genetic recombination. By analyzing the frequency of recombination, Sturtevant could estimate the relative distances between genes on a chromosome. His innovative approach laid the groundwork for subsequent studies on gene organization on chromosomes.
Recent research has further expanded our understanding of chromosome mapping:
- Microsatellite Markers in Mapping: A study highlighted the use of microsatellite markers to localize the Rfv3 gene to a specific region of the mouse chromosome. This technique offers precision in mapping specific genes to their respective locations on chromosomes.
- Comparative Approach in Mapping: Research has described a comparative method for creating physical and linkage maps of genes on canine chromosomes. This approach is particularly beneficial in understanding the genetic makeup of different species and drawing parallels.
- Physical Mapping in Organisms: A novel physical mapping approach has been developed that refines the positions of sex-determining genes on specific chromosomes. This method is adaptable and can be applied to various organisms, showcasing its versatility.
Definition of Chromosome Mapping
Chromosome mapping is the process of determining the relative positions of genes on a chromosome, allowing for the organization and understanding of genetic information.
Types of Chromosome Mapping
Chromosome mapping is a pivotal technique in the realm of genetics, enabling scientists to discern the spatial arrangement of genes and other significant sequences on chromosomes. This mapping is instrumental in understanding genetic diseases, inheritance patterns, and the intricate architecture of the genome. Broadly, chromosome mapping bifurcates into two primary categories: Genetic Mapping and Physical Mapping. Each of these categories possesses distinct methodologies and objectives, which are elucidated below.
- Genetic Mapping:
- Definition: Genetic mapping, often referred to as linkage mapping, is a method that determines the relative positions of genes on a chromosome based on the frequency of their recombination. It is grounded in the principle that genes located in close proximity on a chromosome tend to be inherited together and are less likely to be separated during the recombination process.
- Methodology: This technique employs the use of genetic markers, which are identifiable DNA sequences. By studying the inheritance patterns of these markers in successive generations, scientists can estimate the distance and order of genes relative to each other.
- Applications: Genetic mapping is pivotal in identifying genes associated with specific diseases. It aids in understanding the genetic basis of certain traits and can be instrumental in plant and animal breeding programs to enhance desirable characteristics.
- Physical Mapping:
- Definition: Contrary to genetic mapping which determines relative positions, physical mapping ascertains the exact location of genes on a chromosome. It provides a more detailed and precise representation of the genomic landscape.
- Methodology: Physical mapping techniques involve direct analysis of the DNA molecule. Methods such as Restriction Fragment Length Polymorphism (RFLP), Fluorescence In Situ Hybridization (FISH), and Contig Mapping are commonly employed. These techniques allow for the direct measurement of the DNA sequence’s physical length and its specific location on the chromosome.
- Applications: Physical maps are quintessential in genome sequencing projects. They provide a foundational framework for sequencing large genomes and are instrumental in identifying structural variations in the genome, such as deletions, duplications, and inversions.
1. Genetic Mapping
In the realm of genetics, understanding the spatial arrangement of genes on chromosomes is of paramount importance. Genetic mapping, often termed linkage mapping, serves as a pivotal technique to delineate this arrangement. This methodological approach facilitates the creation of genetic maps, which are essentially representations of the gene positions on chromosomes. These positions are ascertained based on the patterns of recombination observed during the chromosomal crossover process.
Fundamentals of Genetic Mapping:
- Linkage Analysis: At the heart of genetic mapping lies linkage analysis. This process involves studying the recombination frequency between genes to discern their relative positions. A pivotal metric in this analysis is the centimorgan (cM), which denotes a recombination frequency of 1%.
- Recombination Frequency: The propensity for recombination between two genes is directly proportional to their distance on a chromosome. As genes drift further apart, the likelihood of recombination escalates, leading to an augmented recombination frequency. This frequency is quantified by comparing the number of recombinant offspring to the total offspring, and it is articulated as a percentage.
Genetic Linkage and Recombination:
- Mendelian Principles: Mendel’s law of independent assortment posits that allele pairs segregate independently. However, subsequent insights post the rediscovery of Mendel’s work unveiled that certain gene pairs, due to their proximal chromosomal location, were co-inherited. This phenomenon is termed genetic linkage.
- Concept of Linkage: Linkage is a foundational concept in genetic mapping. Genes in close chromosomal proximity are termed linked genes, and their adjacency augments the probability of their concurrent inheritance. However, it’s imperative to note that not all genes on a chromosome exhibit linkage. Those spaced further apart have an elevated chance of separation during the recombination process.
- Recombination Dynamics: Recombination is a hallmark event of meiosis. During this phase, homologous chromosomes congregate to form bivalents. This congregation can instigate crossing-over between chromatids, culminating in the exchange of genetic segments. The propensity for this crossover is inversely proportional to the distance between genes; genes in closer proximity exhibit reduced crossover frequencies.
Genetic Mapping Molecular Markers
In the intricate realm of genetics, molecular markers have emerged as invaluable tools for mapping and understanding the genome’s architecture. These markers, essentially specific DNA sequences, exhibit variations and can be pinpointed using advanced molecular methodologies. Their primary role is to demarcate positions on the genome, thereby facilitating a deeper comprehension of genetic structures and functions.
Fundamentals of Molecular Markers:
- Definition: A molecular marker is a distinct DNA sequence that manifests variation. Its primary utility lies in mapping, where it aids in determining its precise locus on the genome.
- Mapping Process: The inheritance patterns of these markers are meticulously observed across subsequent generations. This observation is typically achieved through techniques such as Southern hybridization and Polymerase Chain Reaction (PCR). The resultant data, which encompasses information on the presence or absence of specific markers, is then processed using specialized software. This software aids in constructing a genetic map, elucidating the relative loci of markers on the genome.
Prominent Molecular Markers in Genetic Mapping:
- Restriction Fragment Length Polymorphisms (RFLPs):
- Overview: RFLPs stand as one of the pioneering molecular markers employed in genetic mapping. They represent distinct patterns of DNA fragments, which are generated by restriction enzymes. These enzymes are adept at recognizing and cleaving DNA at designated sites.
- Functionality: Variations can occur in these restriction sites within genomic DNA. Such variations induce alterations in the restriction patterns, even among genomes that share close genetic ties. By examining the inheritance pattern of RFLPs, their position on a genome can be ascertained.
- Simple Sequence Length Polymorphisms (SSLPs):
- Overview: SSLPs are DNA markers typified by repetitive sequences, with the number of repeats exhibiting variation.
- Types:
- Minisatellites: Often referred to as Variable Number of Tandem Repeats (VNTRs), minisatellites possess longer repeat units, extending up to 25 base pairs.
- Microsatellites: Commonly known as Simple Tandem Repeats (STRs), microsatellites are characterized by shorter repeat units, predominantly dinucleotide or tetranucleotide sequences.
- Single Nucleotide Polymorphisms (SNPs):
- Overview: SNPs represent variations within a genome, characterized by divergent nucleotides at specific loci.
- Detection: Techniques such as oligonucleotide hybridization are employed for SNP detection. This method facilitates selective pairing between an oligonucleotide and its corresponding DNA molecule. Advanced methodologies, including DNA chip technology and solution hybridization, have been devised for SNP screening. These techniques surpass traditional gel-based methods in terms of efficiency and speed.
Limitations of Genetic Mapping
Genetic mapping, while instrumental in elucidating the relative positions of genes on chromosomes, is not without its limitations. This technique, grounded in the study of recombination patterns, offers a broad overview of the genomic landscape but falls short in certain aspects. Herein, we delve into the inherent constraints of genetic mapping and the subsequent implications for genetic research.
- Lack of Physical Distance Information:
- Genetic mapping primarily provides a relative positioning of genes, offering an outline of their arrangement. However, it does not furnish details regarding the actual physical distance between these genes. This absence of precise locational data can pose challenges in understanding the intricate spatial relationships within the genome.
- Variable Recombination Levels:
- The genome is a dynamic entity, with certain regions exhibiting differential rates of recombination. Notably, there exist regions termed ‘recombination hotspots’ that manifest elevated recombination frequencies. Such disparities can skew the accuracy of genetic mapping, potentially leading to misinterpretations.
- Resolution Constraints in Certain Organisms:
- The efficacy of genetic mapping is intrinsically tied to the number of observed crossovers. In organisms where procuring a substantial progeny count is challenging, the resolution of the genetic map becomes inherently limited. This constraint can impede the generation of detailed and comprehensive genetic maps.
- Necessity for Complementary Mapping Techniques:
- Given the aforementioned limitations, it becomes imperative to integrate data derived from alternative mapping methodologies. By amalgamating information from diverse sources, a more holistic and accurate representation of the genome can be achieved.
- Advent of Physical Mapping:
- To circumvent the constraints posed by genetic mapping, researchers have pioneered various physical mapping techniques. These methods aim to provide a detailed and precise representation of the genome, addressing the gaps left by traditional genetic mapping.
2. Physical Mapping
In the intricate domain of genetics, understanding the precise spatial arrangement of DNA sequences on chromosomes is paramount. Physical mapping emerges as a pivotal technique in this context, aiming to delineate the exact loci of DNA sequences on chromosomes. Unlike its counterpart, genetic mapping, which provides a relative positioning of genes, physical mapping offers a direct and detailed representation of the chromosomal architecture.
Core Aspects of Physical Mapping:
- Unit of Measurement:
- Physical mapping employs the base pair as its fundamental unit of measurement. This granularity allows for an accurate depiction of the distance between DNA sequences, ensuring precision in the mapping process.
- Direct Representation:
- One of the salient features of physical mapping is its ability to provide a direct portrayal of the chromosome’s physical structure. This representation elucidates the positions of genes along the DNA sequence, offering insights into the genomic landscape.
- Markers in Physical Mapping:
- Physical mapping leverages a variety of markers to pinpoint DNA sequences on chromosomes. Some of the prominent markers include:
- Expressed Sequence Tags (ESTs): These are short sub-sequences of transcribed DNA, serving as a vital resource to identify gene transcripts in genomic DNA.
- Sequence Tagged Site (STS) Markers: STS markers are unique DNA sequences that can be easily detected and located. Their uniqueness ensures that they occur only once in the genome, making them invaluable in the mapping process.
- Genome-wide DNA Sequences: These sequences encompass the entirety of an organism’s DNA, providing a comprehensive framework for physical mapping.
- Physical mapping leverages a variety of markers to pinpoint DNA sequences on chromosomes. Some of the prominent markers include:
Physical Mapping Methods
Physical mapping stands as a cornerstone in genetic research, offering precise locational information of genes on chromosomes. Several advanced methodologies have been developed to facilitate this intricate process. Herein, we delve into the prominent methods employed in physical mapping, elucidating their mechanisms and applications.
- Cytogenetic Mapping:
- Overview: This method hinges on the unique banding patterns exhibited by chromosomes. When visualized under a microscope, each chromosome manifests a distinct sequence of light and dark bands.
- Mechanism: These bands are systematically numbered, enabling the precise identification of specific chromosomal regions. By correlating gene positions with their corresponding chromosomal bands, cytogenetic maps, or idiograms, are constructed.
- Restriction Mapping:
- Overview: Restriction mapping is centered on pinpointing the loci of restriction sites within a DNA fragment.
- Mechanism: The method involves juxtaposing the sizes of DNA fragments generated by disparate restriction enzymes. While RFLPs serve as markers for genetic mapping, identifying variable restriction sites, many sites remain invariant. To map these non-variable sites, restriction mapping emerges as an alternative.
- Fluorescence in situ Hybridization (FISH):
- Overview: FISH offers a visual representation of specific DNA sequences on chromosomes.
- Mechanism: DNA probes are labeled and subsequently hybridized onto intact chromosomes. While initial FISH methodologies employed radioactive probes, advancements led to the development of fluorescent DNA labels, enhancing sensitivity and resolution.
- Sequence Tagged Site (STS) Mapping:
- Overview: STS mapping is instrumental in crafting detailed physical maps of expansive genomes.
- Mechanism: The process employs short, unique DNA sequences termed STSs, ranging between 100 to 500 base pairs. By identifying fragments containing specific STSs through hybridization analysis or PCR, a map is constructed. Various sources, including expressed sequence tags (ESTs), SSLPs, or random genomic sequences, can yield STSs. Notably, ESTs, derived from partially sequenced clones from a cDNA library, are frequently utilized markers in STS mapping.
- Radiation Hybrid (RH) Mapping:
- Overview: RH mapping is a method designed to gauge the distance between genetic markers.
- Mechanism: The technique employs radiation to fragment DNA. By modulating radiation exposure, DNA breaks between linked markers can be induced, resulting in a more granular map. RH mapping proves invaluable in pinpointing genetic markers and genomic fragments, especially in regions with scarce highly polymorphic markers.
Physical Mapping Limitations
Physical mapping, while instrumental in pinpointing the precise loci of DNA sequences on chromosomes, is not devoid of challenges. This method, which offers a detailed representation of the chromosomal structure, encounters several limitations that can impact its accuracy and comprehensiveness. Herein, we explore the inherent constraints of physical mapping and their implications for genetic research.
- Potential for Incorrect Mapping:
- DNA fragments, during the mapping process, are susceptible to inaccuracies. Factors such as fragment breakage, deletions during replication, or contamination from host genetic material can lead to erroneous mapping. Such inaccuracies can skew the resultant map, potentially leading to misinterpretations.
- Incomplete Coverage:
- One of the salient challenges in physical mapping is the potential for missing or incomplete coverage of DNA fragments. This limitation can result in gaps in the map, rendering it less comprehensive. Such gaps can impede a holistic understanding of the genomic landscape.
- Constraints of Restriction Mapping:
- The restriction mapping method, while effective for certain genomes, encounters challenges when applied to larger genomes. The sheer size and complexity of expansive genomes can render restriction mapping less effective, limiting its applicability.
- Challenges with FISH:
- Fluorescence in situ Hybridization (FISH), though a powerful tool in physical mapping, is fraught with difficulties. The process is intricate, and data accumulation is often protracted. Moreover, a single FISH experiment yields a limited number of map positions, constraining the depth of insights obtained.
- Integration with Genetic Mapping:
- Given the aforementioned limitations, it becomes imperative to amalgamate physical mapping with genetic mapping. By cross-referencing and integrating data from both methodologies, the overall mapping accuracy can be enhanced. This synergistic approach ensures a more comprehensive and accurate representation of the genome.
How to Prepare Chromosome Maps
Chromosome mapping is a pivotal technique in genetics, enabling researchers to determine the relative positions of genes on a chromosome. This methodological approach is based on the principle of recombination frequencies between genes. Here, we elucidate the process of preparing chromosome maps, drawing insights from specific examples.
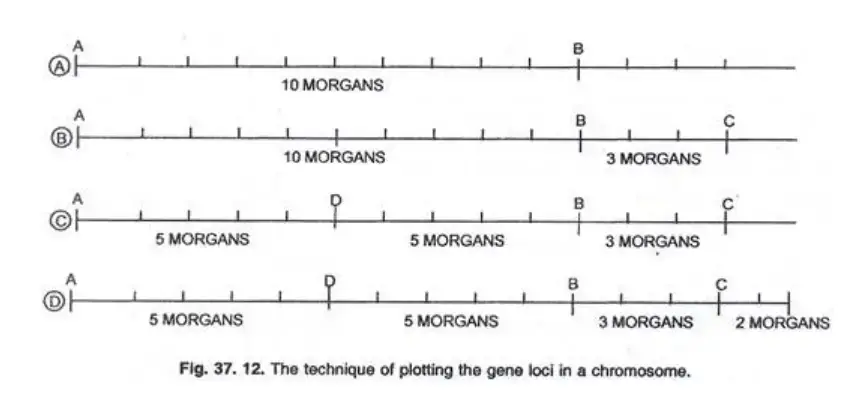
Steps in Preparing Chromosome Maps:
- Determine Recombination Frequencies: Begin by identifying the crossing over percentages between genes. For instance, if genes A and B have a recombination frequency of 10 Morgan’s, they are represented 10 arbitrary units apart on a linear scale.
- Positioning Based on Multiple Genes: To determine the position of a third gene, C, consider its recombination frequencies with both A and B. If A-C has a frequency of 13% and B-C has 3%, then C is plotted 3 Morgan’s to the right of B.
- Continuation of Mapping: Using the aforementioned technique, the positions of subsequent genes, such as D and E, can be plotted. For instance, if A-D and B-D both have a 5% crossover rate, D is plotted equidistant between A and B.
- Finalizing the Map: Once all genes are plotted based on their recombination frequencies, the final chromosome map is constructed, showcasing the relative positions of all genes.
Example:
Consider genes B (black body), cn (cinnabar eye), and vg (vestigial wings) with the following recombination frequencies:
- B-vg: 18%
- B-cn: 9%
- cn-vg: 9.5%
Starting with gene B, vg is plotted 18 Morgan’s away. Given that cn is 9 Morgan’s from B and 9.5 Morgan’s from vg, it is positioned approximately in the middle, resulting in the sequence: b – cn – vg.
The cumulative distance between cn and vg is 18.5 map units, aligning closely with the observed 18 map units for b – vg. This method has facilitated the mapping of numerous mutant genes in organisms like Drosophila.
Applications and Advancements:
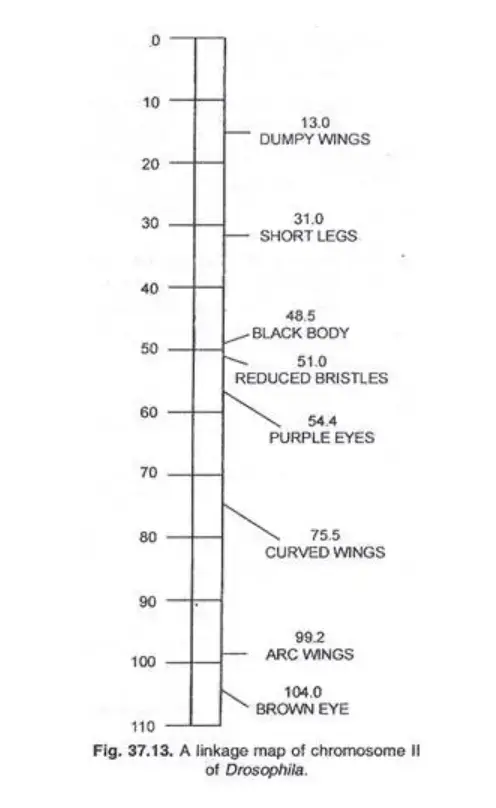
Chromosome maps were initially developed for organisms such as Drosophila, maize, and Neurospora. Modern advancements have expanded this to other organisms, viruses, and even the circular DNA of certain bacteria.
For instance, Drosophila’s chromosome II map showcases the relative positions of its genes, providing invaluable insights into its genetic architecture.
Map Units
In the realm of genetics, map units play a pivotal role in understanding the relative positions of genes on chromosomes. A map unit, often denoted as a centiMorgan (cM), is a unit of measurement that represents the frequency of recombination between genes. Specifically, 1% crossing over between two linked genes is equivalent to 1 map unit or 1 centiMorgan. When the crossing over reaches 100%, it is termed as a Morgan (M), and at 10%, it is referred to as a deciMorgan (dM). The term “centiMorgan” pays homage to T.H. Morgan, a pioneering figure in experimental genetics.
Examples:
Example 1: In Drosophila melanogaster, a series of double crosses were conducted between normal flies and those with recessive genes: sc (scute), ec (echinus), and cv (cross veinless). These genes are sex-linked, residing on the X-chromosome. The observed recombination frequencies were:
- sc and ec: 7.6%
- ec and cv: 9.7%
- sc and cv: 17.3%
From this data, it was deduced that the X-chromosome has the gene sc at one end, followed by gene ec 7.6 cM away, and gene cv 9.7 cM from ec and 17.3 cM from sc. This arrangement confirms that cv is at the opposite end, with ec positioned between sc and cv.
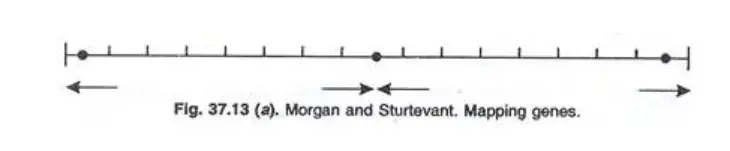
Example 2: Morgan and Sturtevant observed the following recombination frequencies in Drosophila:
- b (Black body) and vg (vestigial wings): 18%
- b and cn (cinnabar eye): 9%
- cn and vg: 9.5%
This data suggests that gene b is at one chromosome end, followed by gene cn 9 cM away, and gene vg 9.5 cM from cn and 18 cM from b. Thus, gene cn is situated between b and vg.
Example 3: Given the map distances:
- A and B: 3 units
- B and C: 10 units
- C and A: 7 units
The arrangement can be deduced as follows: B cannot be on the same side as C since the distance between B and C would then be 4 units (7-3), contrasting the observed 10 cM. Hence, B must be on the opposite side of C, with A positioned between them, 3 units from B and 7 units from C.
Importance of Chromosome Mapping
Chromosome mapping, a pivotal technique in genetics, offers profound insights into the spatial arrangement of genes on chromosomes. This methodological approach not only elucidates the genomic architecture but also plays a crucial role in advancing our understanding of gene interactions, structure, function, and evolutionary patterns. Herein, we delve into the multifaceted importance of chromosome mapping in the realm of genetic research.
- Understanding Genomic Organization:
- Chromosome mapping serves as a foundational tool for deciphering the organization and arrangement of genes on chromosomes. By providing a detailed representation of gene positions, it facilitates a comprehensive understanding of the genomic landscape.
- Elucidating Gene Interactions:
- One of the salient features of chromosome mapping is its ability to identify the precise loci of genes on chromosomes. This precision is instrumental in understanding the intricate interactions between genes, shedding light on their collaborative roles in various biological processes.
- Insights into Gene Structure and Function:
- Beyond mere positioning, chromosome mapping offers insights into the inherent structure and function of genes. By delineating the genomic architecture, it paves the way for a deeper comprehension of how genes operate and interact within the cellular milieu.
- Studying Genetic Disorders:
- Chromosome mapping emerges as an invaluable tool in the study of genetic disorders. By providing information about gene locations, it aids researchers in identifying genes associated with specific disorders. This knowledge is pivotal for diagnostic, therapeutic, and preventive measures in medical genetics.
- Understanding Evolutionary Patterns:
- Chromosome mapping also plays a significant role in evolutionary biology. By comparing subtle variations in gene arrangements among related species, it offers insights into evolutionary trajectories and patterns. Such comparisons elucidate the genetic underpinnings of speciation and evolutionary divergence.
FAQ
What is chromosome mapping?
Chromosome mapping is a technique used to determine the location of genes on a chromosome. It provides a visual representation of the arrangement and sequence of genes.
How does chromosome mapping differ from genetic mapping?
While both methods aim to determine the location of genes, genetic mapping determines the relative positions based on genetic linkage, whereas chromosome mapping provides a more direct representation of the physical locations on a chromosome.
Why is chromosome mapping important?
Chromosome mapping is crucial for understanding the organization of genes on chromosomes, studying genetic disorders, understanding gene interactions, and researching evolutionary patterns among species.
What are the main methods used in chromosome mapping?
Common methods include cytogenetic mapping, restriction mapping, fluorescence in situ hybridization (FISH), sequence tagged site (STS) mapping, and radiation hybrid (RH) mapping.
How does FISH contribute to chromosome mapping?
FISH allows visualization of specific DNA sequences on chromosomes by using fluorescently labeled probes. It provides a direct method to locate and identify gene positions on chromosomes.
What are molecular markers, and why are they important in chromosome mapping?
Molecular markers are specific DNA sequences that show variation and can be identified using molecular techniques. They are crucial for locating positions on the genome and aiding in the mapping process.
How does chromosome mapping aid in the study of genetic disorders?
By determining the exact location of genes associated with specific disorders, chromosome mapping facilitates the study, diagnosis, and potential treatment of genetic diseases.
Can chromosome mapping help in understanding evolution?
Yes, by comparing gene arrangements among related species, chromosome mapping offers insights into evolutionary trajectories, helping to trace the genetic history and divergence of species.
What are the limitations of chromosome mapping?
Some limitations include potential inaccuracies due to DNA fragment breakage, incomplete coverage of DNA fragments, and challenges with certain mapping methods like FISH which can be slow and intricate.
How is chromosome mapping used in modern genetics and biotechnology?
Chromosome mapping is employed in various areas, including gene therapy, drug development, agricultural biotechnology (for crop improvement), and in studying the genetic basis of complex diseases.
References
- National Center for Biotechnology Information. (n.d.). Molecular Biology of the Cell. Retrieved from https://www.ncbi.nlm.nih.gov/books/NBK22266/
- BYJU’S. (n.d.). What is Chromosome Mapping? Retrieved from https://byjus.com/question-answer/what-is-chromosome-mapping/
- Nature Education. (n.d.). Chromosome Mapping & Idiograms. Retrieved from http://www.nature.com/scitable/topicpage/chromosome-mapping-idiograms-302
- International Society of Genetic Genealogy Wiki. (n.d.). Chromosome mapping. Retrieved from https://isogg.org/wiki/Chromosome_mapping
- Ram Jaipal College. (n.d.). [PDF file]. Retrieved from https://online.ramjaipalcollege.org/uploads/files/20200505023720.pdf
- ScienceDirect. (n.d.). Chromosome Map – an overview. Retrieved from https://www.sciencedirect.com/topics/medicine-and-dentistry/chromosome-map
- Vedantu. (n.d.). What is Chromosome Mapping? Retrieved from https://www.vedantu.com/question-answer/what-is-chromosome-mapping-class-11-biology-cbse-605aa590d1abc72e1efaafbd
- Encyclopedia.com. (n.d.). Chromosome Mapping. Retrieved from https://www.encyclopedia.com/science/encyclopedias-almanacs-transcripts-and-maps/chromosome-mapping-0
- ScienceDirect. (n.d.). Chromosome Map – an overview. Retrieved from https://www.sciencedirect.com/topics/biochemistry-genetics-and-molecular-biology/chromosome-map
- Khan Academy. (n.d.). Linkage mapping. Retrieved from https://www.khanacademy.org/science/ap-biology/heredity/non-mendelian-genetics/a/linkage-mapping
- LibreTexts. (n.d.). Gene Mapping and Chromosomal Karyotypes. Retrieved from https://bio.libretexts.org/Bookshelves/Biochemistry/Fundamentals_of_Biochemistry_(Jakubowski_and_Flatt)/03%3A_Unit_III-_Information_Pathway/23%3A_Chromosome_Structure/23.01%3A_Gene_Mapping_and_Chromosomal_Karyotypes
- YourArticleLibrary.com. (n.d.). Chromosome Mapping: Notes on Chromosome Mapping. Retrieved from https://www.yourarticlelibrary.com/biology/chromosome-mapping-notes-on-chromosome-mapping-biology/6599
- Encyclopædia Britannica. (n.d.). Chromosome map. Retrieved from https://www.britannica.com/science/chromosome-map
- Wikipedia. (n.d.). Gene mapping. Retrieved from https://en.wikipedia.org/wiki/Gene_mapping
- Biotechnology Notes. (n.d.). Chromosome Mapping and Its Significance. Retrieved from https://www.biotechnologynotes.com/genetics/chromosome-mapping/chromosome-mapping-and-its-significance-genetics-biotechnology/13352
- Science. (n.d.). [Article]. Retrieved from https://www.science.org/doi/10.1126/science.178.4064.949