What are Cell wall synthesis inhibitors?
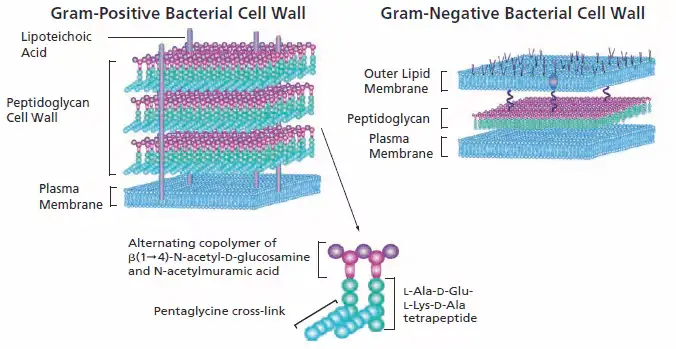
Cell wall synthesis inhibitors encompass antibiotics like β-lactams and glycopeptides. These antibiotics play a crucial role in treating infections by targeting the bacterial cell wall, an essential structure for bacterial survival. Understanding the mechanisms and types of these inhibitors provides insight into their therapeutic applications.
β-Lactam Antibiotics
β-lactam antibiotics include several subclasses: penicillins, cephalosporins, monobactams, and carbapenems.
- Penicillins: This group includes ampicillin and oxacillin. They interfere with the final stages of bacterial cell wall synthesis.
- Cephalosporins: Examples are cefpodoxime and cefepime. These are broad-spectrum antibiotics effective against a variety of bacteria.
- Monobactams: Aztreonam is the only commercially available drug in this class. It is particularly effective against Gram-negative bacteria.
- Carbapenems: This class includes imipenem, meropenem, and ertapenem. These are highly effective against multi-resistant bacterial strains.
Glycopeptide Antibiotics
The primary glycopeptide antibiotics are vancomycin and teicoplanin. They inhibit cell wall synthesis by binding to the D-alanyl-D-alanine termini of peptidoglycan precursors, thus preventing cross-linking and cell wall formation.
Mechanism of Action
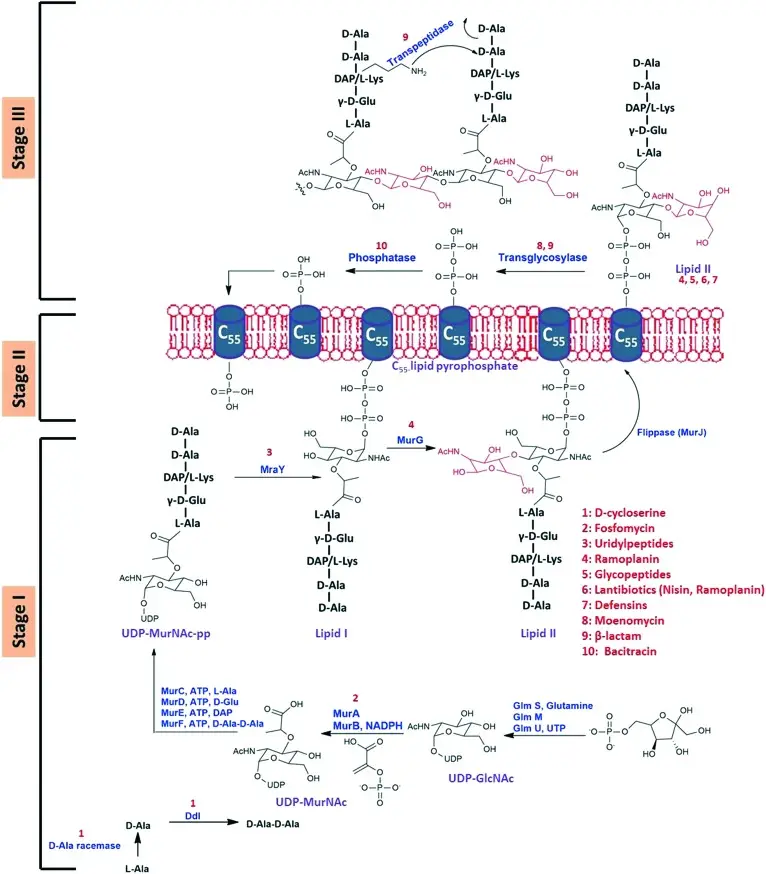
The synthesis of the bacterial cell wall involves a complex process that occurs in three main stages: cytoplasmic, membrane-associated, and periplasmic.
1. Cytoplasmic Stage
In the cytoplasm, the synthesis of peptidoglycan precursors begins. The process starts with UDP-N-acetylglucosamine (UDP-GlcNAc), which undergoes several enzymatic reactions:
- MurA and MurB catalyze the formation of UDP-N-acetylmuramic acid (UDP-MurNAc).
- MurC, MurD, MurE, and MurF enzymes sequentially add amino acids like alanine and glutamic acid to UDP-MurNAc.
- Alanine ligase and alanine racemase are crucial for the formation of the dipeptide D-Ala-D-Ala, a critical component for peptidoglycan cross-linking.
2. Membrane-Associated Stage
In this stage, lipid intermediates are synthesized. These intermediates are critical for transporting peptidoglycan precursors across the cytoplasmic membrane.
3. Periplasmic Stage
The final stage involves the polymerization and cross-linking of peptidoglycan chains. This is facilitated by:
- Glycosyltransferases, which polymerize the glycan strands.
- Transpeptidases, which cross-link these strands to form a rigid cell wall structure. These enzymes include Penicillin-Binding Proteins (PBPs) and RodA proteins.
Key Components in Cell Wall Synthesis
Several components are integral to the process:
- Mur enzymes: These are essential for the initial stages of peptidoglycan precursor synthesis.
- Transpeptidases and glycosylases: These enzymes facilitate the assembly and cross-linking of peptidoglycan.
- Lipid components: These serve as carriers for the peptidoglycan precursors across the membrane.
Process/Steps of Inhibition
The inhibition of bacterial cell wall synthesis involves a series of precise biochemical interactions. These steps disrupt critical processes, leading to bacterial cell death. The main classes of cell wall synthesis inhibitors include beta-lactams and glycopeptides, each targeting different stages of the cell wall formation.
Beta-Lactam Antibiotics
Beta-lactams interfere with the synthesis of the bacterial cell wall by mimicking the D-alanine-D-alanine structure, a key component in peptidoglycan synthesis. This process involves several steps:
- Binding to Transpeptidase Enzymes: Beta-lactams bind to transpeptidase enzymes, also known as penicillin-binding proteins (PBPs). These enzymes are crucial for cross-linking peptidoglycan strands.
- Disruption of Peptide Cross-Linking: The beta-lactam structure interferes with the transpeptidase reaction, preventing the cross-linking of glycine residues in the peptidoglycan chain.
- Formation of Defective Peptidoglycan: Without proper cross-linking, new peptidoglycan chains are weak and defective.
- Cell Rupture: The defective cell wall cannot withstand osmotic pressure, leading to cell rupture and death.
Additionally, beta-lactams affect the autolytic enzymes in bacterial cells. These enzymes, normally involved in cell growth and division, become unregulated in the presence of beta-lactams, further contributing to bacterial cell death.
Glycopeptide Antibiotics
Glycopeptides inhibit cell wall synthesis through a different mechanism, primarily targeting the lipid II component and pentapeptide chains in peptidoglycan synthesis. The steps include:
- Binding to Lipid II: Glycopeptides bind to the lipid II component, which is essential for transporting peptidoglycan precursors across the cell membrane.
- Prevention of Lipid Recycling: This binding prevents the recycling of the central lipid transporter, halting the addition of new peptidoglycan units.
- Inhibition of Transglycosylation and Transpeptidation: Glycopeptides link with pentapeptide chains, inhibiting the enzymatic activities of transglycosylation and transpeptidation. This prevents the synthesis of peptidoglycan, leading to cell death.
Vancomycin, a key glycopeptide antibiotic, exemplifies this process:
- Blocking PBPs: Vancomycin binds to the D-alanyl-D-alanine terminus, preventing penicillin-binding proteins from functioning. This inhibits cell wall synthesis and causes bacterial lysis.
Some glycopeptides also cause permeabilization and depolarization of the bacterial cell membrane. This further disrupts cell wall synthesis and leads to bacterial death.
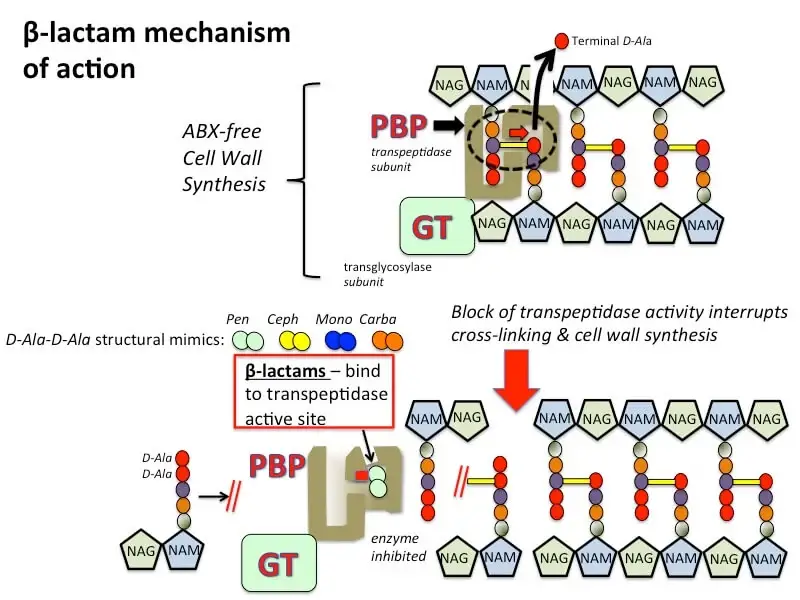
Mechanism of action of beta-lactam antibiotics
Beta-lactam antibiotics are a cornerstone in the treatment of bacterial infections. They function by targeting the bacterial cell wall, a vital component for bacterial survival. This mechanism involves several detailed and sequential steps that disrupt cell wall synthesis.
Key Steps in the Mechanism of Action
- Structural Mimicry: Beta-lactam antibiotics, including penicillins, cephalosporins, monobactams, and carbapenems, share a structural resemblance to the D-alanine-D-alanine dipeptide. This dipeptide is crucial for the bacterial cell wall’s peptidoglycan synthesis.
- Binding to Transpeptidases: The primary target of beta-lactam antibiotics is the transpeptidase enzyme, also known as penicillin-binding proteins (PBPs). These enzymes catalyze the cross-linking of the peptidoglycan strands, which is essential for maintaining the rigidity and integrity of the bacterial cell wall.
- Inhibition of Cross-Linking: In the absence of beta-lactam antibiotics, transpeptidases facilitate the formation of covalent bonds between adjacent glycan chains, involving the removal of a terminal D-alanine residue from one of the peptidoglycan precursors. Glycosyltransferases (GTs), either as separate subunits or associated with transpeptidases, create bonds between N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM) molecules. This results in a robust and protective cell wall.
- Competitive Inhibition: Beta-lactam antibiotics bind tightly to the active site of transpeptidases. This binding is competitive, as beta-lactams mimic the D-alanine-D-alanine substrate. By occupying the active site, beta-lactams prevent the transpeptidase enzyme from catalyzing the cross-linking of peptidoglycan strands.
- Cell Wall Weakening: The inhibition of transpeptidase activity prevents the formation of peptide cross-links, leading to the production of a defective cell wall. Without proper cross-linking, the cell wall is unable to maintain its structural integrity.
- Osmotic Lysis: The weakened cell wall is incapable of withstanding osmotic pressure, resulting in cell lysis and bacterial death. This process is particularly effective against actively growing and dividing bacterial cells.
Detailed Components Involved
- Transpeptidases (PBPs): These enzymes are essential for peptidoglycan cross-linking, providing the bacterial cell wall with rigidity.
- Glycosyltransferases (GTs): These enzymes facilitate the formation of covalent bonds between NAM and NAG, crucial components of the peptidoglycan structure.
- Peptidoglycan Precursors: The building blocks of the bacterial cell wall, including N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM).
Mechanisms of Resistance
The increasing resistance to beta-lactam antibiotics among bacterial pathogens is a significant challenge in medical microbiology. This resistance arises through several mechanisms that allow bacteria to survive despite the presence of these antibiotics.
Key Mechanisms of Resistance
- Alteration of Penicillin-Binding Proteins (PBPs):
- Bacteria can produce altered PBPs that have a low affinity for beta-lactam antibiotics. For instance, the production of PBP2a, encoded by the mecA gene, confers resistance to methicillin and oxacillin in Staphylococcus aureus. This protein can perform transpeptidase activity even in the presence of beta-lactams, allowing the bacterium to continue synthesizing its cell wall.
- Enzymatic Degradation:
- Beta-lactamases are enzymes produced by both Gram-positive and Gram-negative bacteria. These enzymes hydrolyze the beta-lactam ring, rendering the antibiotic ineffective. Gram-positive bacteria typically produce penicillinase, which can mutate to alter its activity, while Gram-negative bacteria express a variety of beta-lactamases, both chromosomally and plasmid-encoded, with broad activity.
- Efflux Pumps:
- Bacteria utilize efflux pumps, which are membrane proteins that expel antibiotics from the cell. These pumps decrease the intracellular concentration of beta-lactams, thereby reducing their effectiveness. Efflux pumps contribute significantly to multidrug resistance (MDR) by expelling a wide range of antibiotics.
- Alteration of Target Sites:
- Resistance to glycopeptides like vancomycin occurs through the alteration of target sites. Bacteria can change the terminal D-alanine-D-alanine of peptidoglycan precursors to D-alanine-D-lactate, which has a reduced affinity for glycopeptides. This modification prevents glycopeptides from binding and inhibiting cell wall synthesis.
- Gene Regulation and Mutations:
- Chromosomal mutations can increase the production of PBP2a, leading to higher resistance levels. These mutations can either directly affect the mecA gene or alter regulatory pathways that increase mecA transcription.
- Plasmid-Mediated Resistance:
- Plasmids can transfer resistance genes between bacteria, spreading resistance traits rapidly. For example, van genes responsible for vancomycin resistance encode dehydrogenases that synthesize lactate, crucial for the formation of unmodified peptidoglycan. Plasmid-encoded beta-lactamases and other resistance factors can be transferred horizontally, enhancing bacterial adaptability.
Specific Examples
- Methicillin-Resistant Staphylococcus aureus (MRSA):
- MRSA expresses PBP2a, which beta-lactams cannot bind. Therefore, beta-lactam antibiotics fail to inactivate the PBP2a, allowing MRSA to resist this subclass of antibiotics.
- Vancomycin-Resistant Enterococci (VRE):
- VRE alters its peptidoglycan precursor from D-alanine-D-alanine to D-alanine-D-lactate, reducing vancomycin binding and conferring resistance.
- Extended-Spectrum Beta-Lactamases (ESBLs):
- ESBL-producing Gram-negative bacteria can hydrolyze a wide range of beta-lactams, including penicillins and cephalosporins, posing a significant treatment challenge.
Examples of Cell wall synthesis inhibitors
Cell wall synthesis inhibitors are a critical class of antibiotics used to treat a variety of bacterial infections. They disrupt the formation of the bacterial cell wall, leading to cell lysis and death. These inhibitors include several subclasses, each with specific examples and applications.
1. Beta-Lactam Antibiotics
Beta-lactam antibiotics are characterized by their beta-lactam ring, which is crucial for their antibacterial activity. They are divided into four main groups: penicillins, cephalosporins, monobactams, and carbapenems.
Penicillins:
- Ampicillin: Effective against a broad range of bacteria, used for respiratory, urinary, and gastrointestinal infections.
- Amoxicillin: Similar to ampicillin but with better oral absorption, used for otitis media, sinusitis, and bronchitis.
- Piperacillin: Often combined with tazobactam, used for more severe infections including hospital-acquired infections.
- Oxacillin: Resistant to beta-lactamase enzymes, used for penicillin-resistant Staphylococcus infections.
Cephalosporins:
- First-generation:
- Cefazolin: Used for surgical prophylaxis and skin infections.
- Cefalexin: Used for respiratory and skin infections.
- Second-generation:
- Cefoxitin: Effective against anaerobic bacteria, used for intra-abdominal infections.
- Cefuroxime: Used for respiratory tract infections and Lyme disease.
- Third-generation:
- Cefixime: Effective against Gram-negative bacteria, used for respiratory and urinary tract infections.
- Ceftazidime: Used for Pseudomonas infections.
- Ceftriaxone: Broad-spectrum, used for severe infections including meningitis.
- Fourth-generation:
- Cefepime: Broad-spectrum, used for severe hospital-acquired infections.
Monobactams:
- Aztreonam: The only commercially available monobactam, effective against Gram-negative bacteria, including Pseudomonas aeruginosa.
Carbapenems:
- Imipenem: Broad-spectrum, used for severe or multi-resistant infections.
- Meropenem: Similar to imipenem, often preferred due to fewer side effects.
2. Glycopeptides
Glycopeptide antibiotics are vital for treating Gram-positive infections, especially those resistant to beta-lactams.
Examples include:
- Vancomycin: Used for serious Gram-positive infections, including MRSA.
- Oritavancin: Long-acting, used for skin and soft tissue infections.
- Teicoplanin: Similar to vancomycin, with a longer half-life.
- Telavancin: Used for complicated skin and soft tissue infections.
- Bleomycin: Primarily used in cancer therapy but has antibacterial properties.
- Ramoplanin: Effective against Gram-positive bacteria, including C. difficile.
- Decaplanin: Investigational drug with potential for Gram-positive infections.
| Class | Subgroup | Examples | Applications |
|---|---|---|---|
| Beta-Lactam Antibiotics | Penicillins | Ampicillin, Amoxicillin, Piperacillin, Oxacillin | Respiratory, urinary, gastrointestinal infections, penicillin-resistant Staphylococcus infections |
| Cephalosporins | First-generation: Cefazolin, Cefalexin | Surgical prophylaxis, skin infections, respiratory infections | |
| Second-generation: Cefoxitin, Cefuroxime | Intra-abdominal infections, respiratory tract infections, Lyme disease | ||
| Third-generation: Cefixime, Ceftazidime, Ceftriaxone | Respiratory, urinary tract infections, Pseudomonas infections, severe infections including meningitis | ||
| Fourth-generation: Cefepime | Severe hospital-acquired infections | ||
| Monobactams | Aztreonam | Gram-negative bacteria, including Pseudomonas aeruginosa | |
| Carbapenems | Imipenem, Meropenem | Severe or multi-resistant infections | |
| Glycopeptides | Vancomycin, Oritavancin, Teicoplanin, Telavancin, Bleomycin, Ramoplanin, Decaplanin | Serious Gram-positive infections, including MRSA, skin and soft tissue infections, cancer therapy (Bleomycin) |
References
- Ghooi RB, Thatte SM. Inhibition of cell wall synthesis–is this the mechanism of action of penicillins? Med Hypotheses. 1995 Feb;44(2):127-31. doi: 10.1016/0306-9877(95)90085-3. PMID: 7596307.
- Sarkar P, Yarlagadda V, Ghosh C, Haldar J. A review on cell wall synthesis inhibitors with an emphasis on glycopeptide antibiotics. Medchemcomm. 2017 Jan 26;8(3):516-533. doi: 10.1039/c6md00585c. PMID: 30108769; PMCID: PMC6072328.
- Bugg, T. D., & Walsh, C. T. (1992). Intracellular steps of bacterial cell wall peptidoglycan biosynthesis: enzymology, antibiotics, and antibiotic resistance. Natural Product Reports, 9(3), 199-215.
- Silver, L. L. (2003). Novel inhibitors of bacterial cell wall synthesis. Current Opinion in Microbiology, 6(5), 431-438.
- Lovering, A. L., Safadi, S. S., & Strynadka, N. C. (2012). Structural perspective of peptidoglycan biosynthesis and assembly. Annual Review of Biochemistry, 81, 451-478.
- Kohanski, M. A., Dwyer, D. J., & Collins, J. J. (2010). How antibiotics kill bacteria: from targets to networks. Nature Reviews Microbiology, 8(6), 423-435.
- Schneider, T., & Sahl, H. G. (2010). An oldie but a goodie–cell wall biosynthesis as antibiotic target pathway. International Journal of Medical Microbiology, 300(2-3), 161-169.
- Typas, A., Banzhaf, M., Gross, C. A., & Vollmer, W. (2012). From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nature Reviews Microbiology, 10(2), 123-136.
- https://accesspharmacy.mhmedical.com/content.aspx?bookid=514§ionid=41817562
- https://www.dentalcare.com/en-us/ce-courses/ce475/inhibitors-of-bacterial-cell-wall-synthesis
- https://www.osmosis.org/learn/Miscellaneous_cell_wall_synthesis_inhibitors
- https://www.creative-biolabs.com/drug-discovery/therapeutics/inhibitors-of-cell-wall-biosynthesis.htm
- https://www.sigmaaldrich.com/IN/en/technical-documents/technical-article/clinical-testing-and-diagnostics-manufacturing/bacteriology/how-antibiotics-work
- https://www.kgmu.org/download/virtualclass/Pharmacology/31.ppt
- https://www.biomol.com/resources/biomol-blog/how-do-antibiotics-affect-cell-wall-synthesis