Human pathogenic bacteria transmitted via the fecal-oral route, i.e., primarily intestinal pathogens, constitute one component of drinkable water quality analysis. Screening water for faecal contamination by testing for the presence of an indicator microbe is more practicable than doing thorough routine examinations for the presence of every type of disease. If you want to know how contaminated your water is, look for indicator microorganisms, which have two key characteristics:
- They normally wouldn’t be found in water and
- Their density in the water should be proportionate to the degree of pollution. Using Escherichia coli as an indicator microbe to identify harmful germs via the fecal-oral pathway was first proposed in the 1890s.
The research done by Theodore Escherich in the 1880s led to the selection of this bacteria. Escherich discovered that Bacillus coli, currently known as E. coli, is found in the intestines (i.e., an enteric bacterium) and faeces of animals and so fits the properties of the indicator microorganism stated above. There are still certain regulations in place that prioritise the absence of coliforms (a group of bacteria that includes E. coli) over other water quality indicators.
The presence of these indicator microorganisms can be detected using a wide variety of methods. The following characteristics are desirable in such methods: The method must be sensitive enough to pick up the indicator even at trace amounts. The method must be able to handle substantial quantities of water. A good method for detecting the existence of the indication should not be complicated, should not cost too much, and should work quickly. By filtering the water via a membrane made of cellulose nitrate or cellulose acetate, Goetz and Tsuneishi were able to isolate and study any bacteria present in the water sample. This method is still in use today.
Purpose
- Multiple water samples from various locations are analysed using the membrane filtration method. In this step, the membrane is incubated on agar. Colonies of bacteria (and other cells) that become trapped on the membrane can be quantified and used to determine the bacterial density of the water samples.
Principle
Total Coliforms & Fecal Coliforms
Membrane filtration can identify total coliforms, which are indicator bacteria. Total coliforms are members of the family Enterobacteriaceae, albeit this classification is more operational than phylogenetic. The term “coliforms” is often used to refer to other types of bacteria besides those found in faeces. Moreover, different countries and public health organisations have different definitions of what constitutes a “total coliform”. A bacteria must meet the following criteria in order to be labelled as “total coliform”:
- Gram-negative rod;
- aerobe or facultative anaerobe;
- non-spore forming;
- and able to ferment lactose into acid in 24 hours at 35oC (if the membrane filtering method is used) or acid and gas in 48 hours at 35o C. (for multiple-tube fermentation technique, not described in this protocol).
Escherichia coli, Enterobacter cloacae, Klebsiella spp., Citrobacter spp., and Serratia marcescens are all examples of coliform bacteria. The vast majority of total coliforms are not dangerous. Fecal coliforms, which live in the intestines and are eliminated in faeces, are a subset of total coliforms. There is a stronger link between the presence of these indicator bacteria and the presence of faecal contamination. Within 24 hours at 44.5o C, this group is able to ferment lactose, resulting in acid (and gas, depending on the method) generation. Due to their ability to thrive in warmer environments, these coliforms are also classified as thermotolerant coliforms. When it comes to faeces, not all coliforms are created equal. Faecal coliforms are often thought of as bacteria from the genera Escherichia and Enterobacter.
Membrane Filtration
Multiple water samples from various locations are analysed using the membrane filtration method. A suitable amount of the material is filtered through a 0.45 mm-pore-size membrane. In this step, the membrane is incubated on agar. Colonies of bacteria (or other cells) that become stuck on the membrane can be quantified and used to determine the bacterial density. If you want to check a water sample for indicator microorganisms using a membrane filtering method, you’ll need to filter it a certain number of times, and those numbers may vary depending on where the sample came from.
| Source | 100ml | 50ml | 10ml | 1ml | 0.1ml | 0.01ml | 0.001ml | 0.0001ml |
| Drinking water | X | |||||||
| Swimming pool | X | |||||||
| Wells, springs | X | X | X | |||||
| Lakes, reservoirs | X | X | X | |||||
| Water supply intakes | X | X | X | |||||
| Bathing beaches | X | X | X | |||||
| River water | X | X | X | X | ||||
| Chlorinated sewage | X | X | X | |||||
| Raw sewage | X | X | X | X |
| Source | 100ml | 50ml | 10ml | 1ml | 0.1ml | 0.01ml | 0.001ml | 0.0001ml |
| Lakes, reservoirs | X | X | ||||||
| Wells, springs | X | X | ||||||
| Water supply intakes | X | X | X | |||||
| Natural Bathing waters | X | X | X | |||||
| Sewage treatment plant | X | X | X | |||||
| Farm ponds, rivers | X | X | X | |||||
| Stormwater runoff | X | X | X | |||||
| Raw sewage | X | X | X | |||||
| Feedlot runoff | X | X | X | |||||
| Sewage sludge | X | X | X |
Water for human consumption should be filtered in duplicate, and all other sample sources should be filtered in triplicate, or three separate amounts. Since membrane filtration is very reproducible, it can be used to detect a wide variety of organisms when combined with the right medium. Since enormous amounts of sample can be filtered, it has the potential to have an extremely low detection limit. However, the volume of material that is manageable to filter may be constrained by the turbidity of the sample. An underestimating of coliform density may occur if the test is impeded by a high concentration of background microorganisms or harmful chemicals.
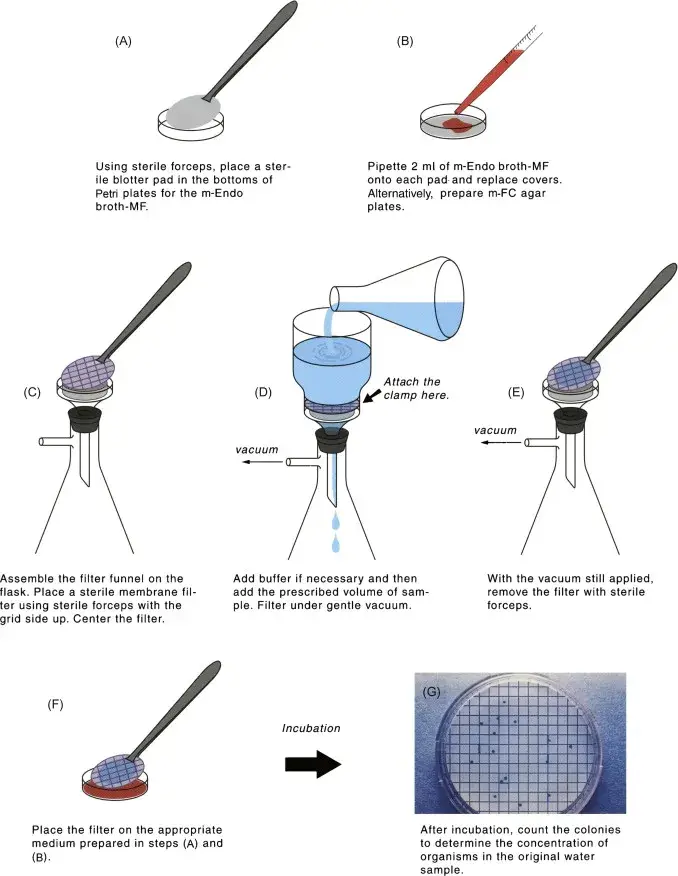
Detection of total coliforms and fecal coliforms
Culture media and incubation conditions selective for Gram-negative rods and/or differential for lactose fermenters make it simple to test for the traits that identify the total coliform and faecal coliform groups in the lab. Some of the most common types of growth media are:
1. Detection of total coliforms – mEndo agar LES
This growing medium includes lactose and a pH indicator whose colour changes when acid is created (from lactose fermentation). Due to the substantial synthesis of aldehydes and acid during lactose fermentation, coliforms generally have a metallic (golden) sheen. These uncommon complete coliform colonies may also be dark red, mucoid, or have a dark centre but lack a metallic sheen. Colonies of E. coli will have a metallic sheen. Importantly, some non-coliforms may exhibit red colonies. Filters used to identify the presence of total coliforms in a water sample should be incubated for 22 to 24 hours at 35 degrees Celsius.
2. Detection of fecal coliforms – mFC agar
This medium for growth contains bile salts. Other bacteria cannot grow in the presence of bile salts, but enteric bacteria can. mFC agar also contains rosolic acid, which inhibits non-fecal coliform bacteria. Aniline blue, a pH indicator, turns dark blue in the presence of acid, hence facilitating the identification of lactose-fermenting bacteria. On this medium, coliforms from faeces create blue colonies, while E. coli forms flat, dark blue colonies. Although rosolic acid is often utilised as a component of mFC agar, it has been argued that it is not essential to this medium. Filters used to identify the presence of faecal coliforms in a water sample should be incubated for 22 to 26 hours at 44.5 degrees Celsius.
For these experiments, either a liquid broth or a solid media may be utilised. This protocol specifies the use of solid media, which is prevalent. It is customary to report total and faecal coliforms in water samples using colony-forming units (CFU) per 100 ml of sample.
Requirement
Nitrocellulose membrane
- 47mm membrane filters with 0.45 mm hole size.
- Nitrocellulose membrane is used to detect total coliform and faecal coliform bacteria in water samples. It has a 47 mm diameter and 0.45 µm pore size.
- During filtration, the narrow hole size of the membrane filter will collect bacterial organisms present in the water sample. The membrane is printed with a grid to facilitate colony counts following incubation.
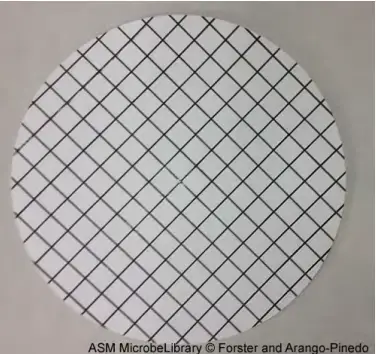
Membrane Filtration Device
- Water can be filtered using either a Buchner funnel or a filtration column. Membrane-filtration column of filtration This filter column can be utilised to identify total coliform and faecal coliform bacteria in water samples.
- First, a vacuum pump is connected to the column. There is a nitrocellulose membrane between the chamber and the catchment vessels. The water sample is subsequently introduced into the column’s chamber.

Vacuum pump
- Preparation of vacuum for membrane filtration. This diagram depicts the assembled filtration column with filter paper and vacuum pump.
- When the vacuum is activated, water will filter through the nitrocellulose membrane and into the collection vessel. The nitrocellulose membrane will capture any bacteria that are present in the water.

Media
The mEndo Agar LES and mFC agar formulas are offered for reference purposes. However, dehydrated media prepared commercially is accessible from a variety of sources. It is strongly advised to use these prepared media according to the directions provided by the manufacturer.
1. mENDO agar (DIFCO) Composition
| Lactose | 9.4g |
| Tryptose | 7.5g |
| Casitone | 7g |
| Thiopeptone | 3.7g |
| Sodium Chloride | 3.7g |
| Potassium Phosphate, dibasic | 3.3 g |
| Sodium Sulfite | 1.6 g |
| Yeast Extract | 1.2 g |
| Potassium Phosphate, monobasic | 1.0 g |
| Basic Fuchsin | 0.8 g |
| SodiumDeoxycholate | 0.1 g |
| Sodium Lauryl Sulfate | 0.05 g |
| Agar | 15 g |
- Suspend components in 1 litre of distilled water containing 20 mL of ethanol at a concentration of 95%.
- Blend thoroughly To completely dissolve the powder, heat while stirring often and bring to a boil; remove from heat immediately.
- DO NOT AUTOCLAVE.
- Let cool to a temperature just above the point at which the agar begins to solidify (45 to 50oC).
- Pour approximately 4 to 6 mL aseptically into 50 mm petri dishes that will accommodate nitrocellulose paper.
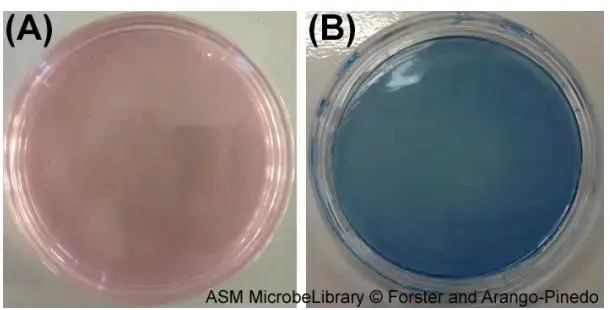
Prior to incubation, mENDO agar (A) and mFC agar (B) are shown. These growth media are selective and differential media used, respectively, for the detection of total coliforms and faecal coliforms. On mENDO agar, coliforms will produce metallic-looking red colonies. Fecal coliforms will produce dark blue colonies on mFC medium.
2. mFC agar – (DIFCO)
| Tryptose | 10.0 g |
| Proteose Peptone No. 3 | 5.0 g |
| Yeast Extract | 3.0 g |
| Lactose | 12.5g |
| Bile Salts No. 3 | 1.5 g |
| Sodium Chloride | 5.0 g |
| Agar | 15.0 g |
| Aniline Blue | 0.1 g |
- Suspend components in 1 litre of distilled water. Blend thoroughly While stirring frequently, bring the liquid to a boil for one minute. Remove from heat right away.
- Add 10 ml of a 1% rosolic acid solution in 0.2N NaOH.
- Continue to heat for another minute. DO NOT AUTOCLAVE.
- Using 1N HCl, adjust the pH to 7.4 if necessary.
- Let cool to a temperature just above the point at which the agar begins to solidify (45 to 50oC).
- Pour approximately 4 to 6 mL aseptically into 50 mm petri dishes that will accommodate nitrocellulose paper.
Forceps, Ethanol, Bunsen Burner
When placing the membrane on the filtration column or the agar plates, these materials should be utilised for aseptic method.
Sterile water (in squirt bottle)
Before adding the water sample to be examined, sterile water is used to ensure that the vacuum chamber is properly adjusted and to remove bacteria and sample residues from the walls of the filter tower or funnel.
Positive Control Sample
100 ml of water containing 5 ml of a 1:100 dilution of an overnight E. coli culture. This sample is to be used as a positive control to ensure that the membrane filtration technology can detect E. coli in a water sample.
Negative Control Sample
Sterile Water
This sample will serve as a negative control when membrane filtration is performed.
Water samples for analysis
Table 1 lists the quantities of water required for testing.
Consumables
- Formaldehyde gas filtration equipment disinfected with methanol (unnecessary in the laboratory, but essential if analyses are done in the field). Methanol is a necessary component. You can’t use vodka or rum in place of ethanol or Methanol.
- Complete with absorbent pads, these membrane filters have a pore size of 0.45 µm and a diameter that is suitable for the filtration equipment being used.
- Chemical cleaner for the lab, and a trash can for used pipettes.
- Media of culture (mEndo Agar LES and mFC Agar).
- Diluted with phosphate-buffered water.
- Culture dishes, either disposable plastic or glass or aluminium (reusable) (disposable).
- If you plan on using a dry incubator, you’ll need polyethylene bags to store your Petri dishes in.
- A magnifying glass (as an aid to counting colonies after filters are incubated).
- Pencils made of wax, used for writing on Petri dish labels.
- Boil-in-the-bag tape.
- glassware and laboratory equipment cleaning detergent.
- Exemplary Positive Control (100 ml of water with a 5 ml of a 1:100 dilution of an overnight culture of E.coli)
- Example of a negative control (sterile water)
- To be examined in water samples: Measurement of the required quantity of water for laboratory experiments.
Procedure
Utilize aseptic process throughout the entire treatment. Prior to use, a Buchner funnel or filtration column must be autoclaved.
Day 1
- Determine the volume of water to be filtered (or the dilution to be created) based on Table 1 and/or 2. Compute and record every pertinent detail (which are the dilutions to be made, what are the volumes of each dilution to be filtered). Before obtaining an aliquot, if necessary, prepare the dilutions and gently mix the sample (or dilution) by inverting the tube or container several times.
Construct the filtration device.- Insert the stopper of a Buchner funnel (with cover) into a flask with a side arm. Ensure that the vacuum pump’s tubing is placed into the flask’s side arm.
- If utilising a filter column, ensure that the vacuum pump’s tubing is placed into the flask’s side arm.
- Turn on Bunsen burner. Pass forceps dipped in ethanol across a flame. This disinfects your forceps.
- a. Remove the cover if using a Buchner funnel (with cover). Take a sterile filter membrane and place it on the filter holder of each funnel using sterile forceps.
- b. If utilising a filter column, remove the upper chamber. Place a sterile filter membrane on top of the catchment vessel using sterile forceps.
- Using the squirt bottle, pour a little amount of sterile water into the filtering apparatus, turn on the pump, and apply vacuum for a few seconds in order to moisten the membrane. Always replace the column’s lid before applying vacuum!
- If the volume to be filtered is modest (less than 10 ml), add 10-20 ml of sterile water to the filtration tower.
- Starting with the smallest volume (or the highest dilution), add the sample volume to be filtered. Before getting the volume, mix the sample (or dilution) by inverting the tube or bottle several times.
- Turn on the pump and transfer the filtrated sample to the flask. Be cautious not to allow the flask or catchment chamber of the column to fill with water; otherwise, the water could be sucked into the pump, causing damage to the vacuum system.
- Using a spray bottle filled with sterile water, clean the tower’s walls. Turn on the vacuum to filter water.
- Transfer the filter membrane to a correctly labelled mENDO agard LES plate using sterile forceps (step 3), preventing the formation of air pockets between the membrane and agar.
- Repeat steps 3 through 8 with the same volume (or dilution).
Repeat the procedure twice more with the same volume, but this time place the membranes on mFC plates. - Repeat steps 3-10 for the following volume (or dilution).
- Repeat steps 3 through 10 with the maximum volume (or lowest dilution)
- Incubate mENDO agar LES plates at 35 ± 0.5 o C for 22 to 24 hours, and mFC plates at 44.5 ± 0.2o C for 22 to 26 hours, all with the lids facing down. In order to regulate the temperature within such a limited range, the mFC agar plates are commonly incubated in a water bath. These plates are placed in watertight plastic bags before being submerged in a water bath.
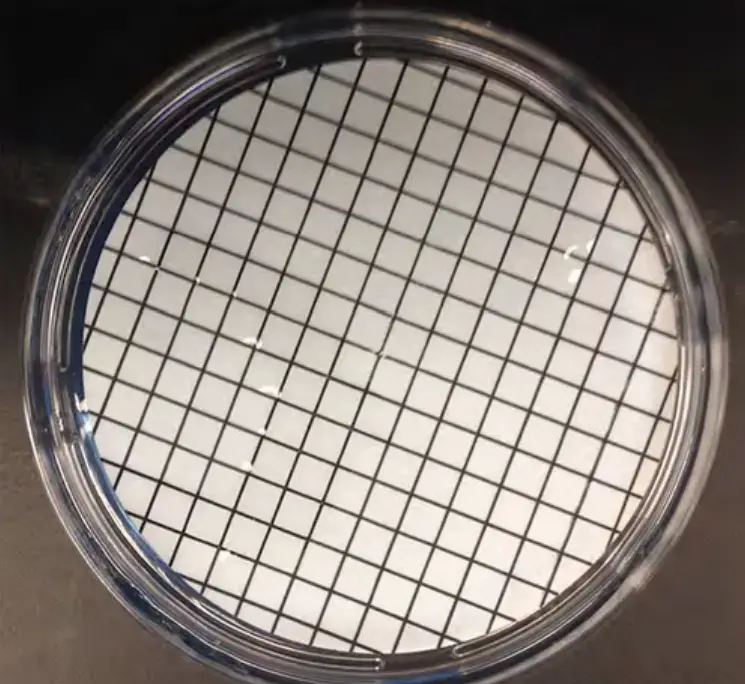
Following vacuum filtration, membrane is positioned on agar. After filtering the water sample using the filtration column, the filter is transferred aseptically, grid-side up, to the plate. The grid should be lightly placed onto the agar so that the nutrient agar is absorbed by the membrane, allowing bacterial colonies to form.
Day 2 (24 hours later)
- After 22 to 24 hours, remove the mEndo agar LES plates from the 35°C incubator and count the colonies that are dark red, mucoid, have a dark core, or (more frequently) create a metallic sheen. Considered to be complete coliform colonies.

- From the mEndo agar LES plates, choose two isolated total coliform colonies and do a gramme stain to determine that they are Gram-negative rods. Use phase-contrast microscopy or the Schaffer-Fulton dye to verify that these colonies contain non-spore-forming bacteria.
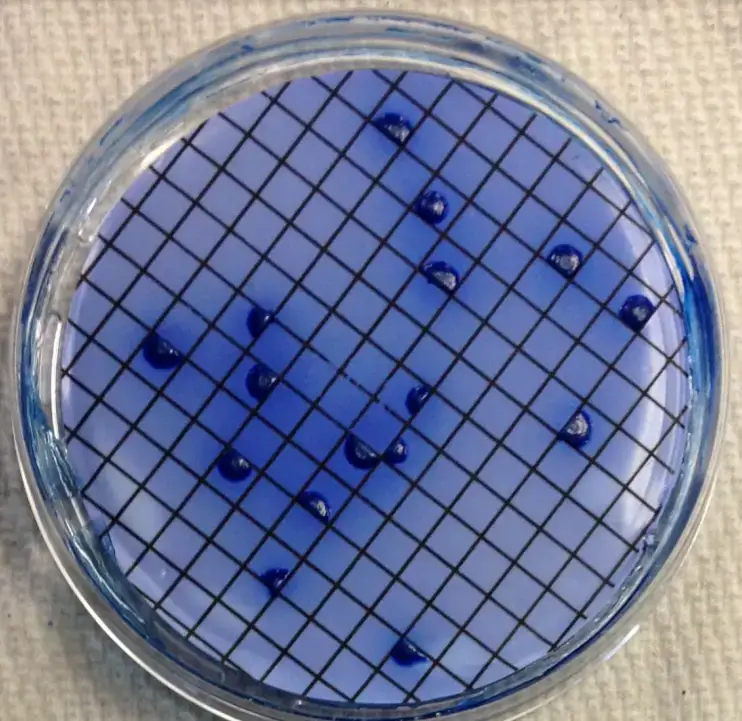
- After 22 to 26 hours, remove the mFC agar plates from the incubator at 44.5o C and count any blue-colored colonies. These are termed coliform faeces colonies.
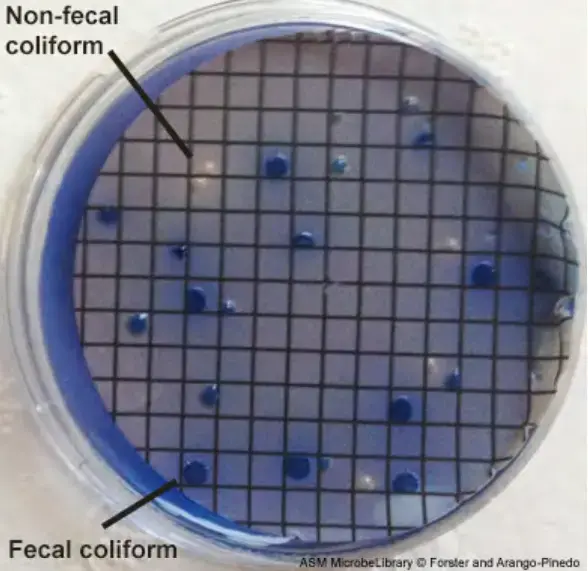
- Choose two faecal coliform colonies isolated on the membrane from the mFC agar plates. Confirm that the organisms are Gram-negative rods that do not produce spores.
- Select two non-total coliform colonies from the LES mEndo agar plates and two non-fecal coliform colonies from the mFC agar plates. These colonies’ bacteria may or may not be Gram-negative.
- Calculate the number of total and faecal coliform CFU per 100 ml for each sample (as described below).
Calculations
Calculating the cell density of the original sample:
- The original density is approximated based on the amount of filtered sample (or the volume of dilution and the dilution factor) and the number of colonies enumerated on the membrane. As counts are provided per 100 ml of sample rather than per ml, the per-ml numbers must be multiplied by 100.
- The best range for calculations is between 20 and 80 total coliforms (with no more than 200 total colonies) each plate. For calculations, counts between 20 and 60 faecal coliforms per plate are optimum.
To calculate density (CFU per 100 ml):
Density = number of colonies on membrane x 100/volume (ml) of undiluted sample filtered
Or if the sample was diluted and a volume of the dilution was filtered:
Original Density = number of colonies on membrane x 100/(volume (ml) filtered x dilution)
Uses of Membrane Filter (MF) Technique
- Samples of water are taken both upstream and downstream from a sewage treatment plant’s outlet, and then diluted serially for analysis.
- Water samples are routinely analysed before being released into a country’s waterways, and EPA-approved guidelines for the determination of faecal contaminating organisms (EPA Method 1103.1) are commonly used for this purpose.
- The safety of the water is measured by the number of coliform bacteria it contains.
- Media components such as serum, certain carbohydrate solutions, certain antibiotics, and other heat-labile substances can be sterilised via membrane filtration rather than being subjected to steam sterilisation in an autoclave.
- Process water testing for Pseudomonas species is common practise in the pharmaceutical and cosmetics industries.
Limitations of Membrane Filter (MF) Technique
- Particulate matter clogs the pores and inhibits passage of the specific volume of water during the processing of turbid specimens that contain large quantities of suspended materials.
- It is important to dilute a portion of the sample in sterile diluent to guarantee that there is adequate volume to filter across the whole surface of the membrane when testing small volumes of sample (such as sewage effluent or of badly polluted surface water).
Precautions
- It is recommended to execute the dilutions (step 1). Otherwise, it may be difficult to identify the precise coliform cell density in water samples.
- It is straightforward to demonstrate that some bacteria in water can, in fact, pass through a 0.45 µm pore-size filter. Simply remove 5 to 10 mL of filtrate from the collection vessel and inoculate a flask or test tube with a liquid growth medium for general purposes, such as tryptic soy broth.
- At 35 °C, incubate the infected material for 24 to 48 hours. Small but viable bacterial cells will have gone through the filter, as shown by the turbidity of the growing medium.
- Typically, 0.22 µm-sized filters are used to “filter-sterilize” water samples. It is also possible to compare the removal capabilities of these two filter types.
References
- Water Quality Monitoring – A Practical Guide to the Design and Implementation of Freshwater Quality Studies and Monitoring Programmes (WHO)
- Brian Forster, Catalina Arango Pinedo. 2015. Bacteriological examination of waters: membrane filtration protocol. American Society for Microbiology (ASM)
- Manual for Bacteriological Analysis of Natural Water Supply Sources in Disaster Situations. Pan American Health Organization (PAHO)
- Pelczar MJ, Chan ECS, Krieg NR (2007). Microbiology. 5th edn. Tata McGraw-Hill.
- https://www.pall.com/en/laboratory/microbiology-qc/laboratory-membrane-filter-technique.html
- https://www.membrane-solutions.com/News_80.htm
- https://www.sciencedirect.com/science/article/pii/S0580951708701387
- http://www.who.int/water_sanitation_health/resourcesquality/wqmchap10.pdf
- http://www.labdepotinc.com/admin/uploads/9222_a.pdf
- http://europepmc.org/backend/ptpmcrender.fcgi?accid=PMC547049&blobtype=pdf
- https://aem.asm.org/content/aem/34/1/42.full.pdf