What is agarose gel electrophoresis?
- Agarose gel electrophoresis (AGE) is an approach that is used to distinguish DNA from RNA based on their molecular sizes.
- The separation of RNA and DNA molecules is accomplished when nucleic acids that are negatively charged travel through an agarose structure under an influence of an electrical fields (electrophoresis).
- In Agarose Gel Electrophoresis the Smaller molecules are more efficient and travel further than larger ones.
- This procedure is easy and quick to execute and can resolve DNA fragments that cannot be separated with other techniques like density gradient centrifugation.
- The DNA’s position in the agarose gel is revealed by staining the gel with a low amount of intercalating fluorescent dyes like Ethidium bromide.
- The procedure consists of three essential steps: Preparation the agarose gel, Electrophoresis of DNA fragments visualization of DNA fragments.
- Agarose gels can be utilized to efficiently separate fragments ranging from 50 bp to a few thousand bases long by altering the porosity the gel and the application of current.
- DNA can be purified from gels by a number of methods such as: Electroelution, electrophoresis onto DEAE Cellulose/Nitrocellulose (NA45) paper, using b-Agarase (from low melting agarose) or using glass beads/ silica etc.
Definition of Agarose Gel Electrophoresis
Agarose gel electrophoresis is a common laboratory technique used to separate and analyze DNA fragments and other biomolecules based on their size. It is widely employed in molecular biology, genetics, and biochemistry research.
The process involves the use of a gel made from agarose, a polysaccharide derived from seaweed. Agarose forms a porous matrix when mixed with a buffer solution and allowed to solidify. This gel matrix creates a molecular sieve through which the biomolecules can migrate under the influence of an electric field.
Aim and Objectives of gel electrophoresis
- Identification of Biomolecules: One of the primary aims of gel electrophoresis is to identify and separate different biomolecules such as DNA, RNA, and proteins. By subjecting these biomolecules to an electrical field, they migrate through the gel matrix based on their size, charge, and shape, allowing for their visualization and analysis.
- Size Determination: Gel electrophoresis is commonly used to determine the size or molecular weight of biomolecules. By comparing the migration distances of unknown samples with known standards of known sizes, researchers can estimate the size of the biomolecules of interest.
- Electrophoretic Mobility: Gel electrophoresis enables the movement of biomolecules under the influence of an electrical current. By applying a voltage across the gel matrix, charged biomolecules migrate towards the oppositely charged electrode, allowing for their separation based on their charge-to-mass ratio.
- Size Estimation: Gel electrophoresis provides an estimation of the size of biomolecules. By comparing the migration distances of different biomolecules on the gel, researchers can make relative assessments of their sizes. Although gel electrophoresis does not provide precise measurements, it offers valuable information about the size distribution and relative size differences among biomolecules.
Agarose gel electrophoresis principle
- Agarose is an amorphous polymer that has been extracted from seaweeds.
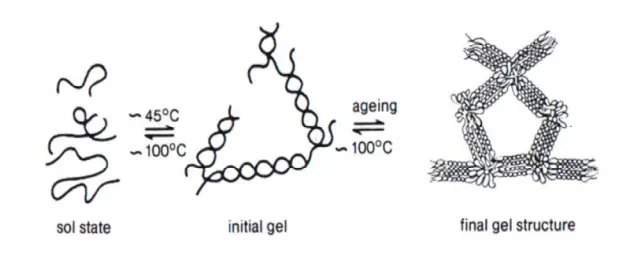
- Pure agarose is that is insoluble in buffer or water at room temperature, but it dissolves upon boiling. The solution that is molten is then poured into a mould and then allowed to set. When it cools, agarose is subjected to polymerization i.e. sugar polymers cross-link each other , causing the solution to gel. The density or the size of the pore that is determined by the the concentration of the agarose.
- DNA’s charge is negative when it’s at neutral pH. When an electrical field is applied to the gel, it is moved toward the anode. The movement of DNA across the gel depends on the size of DNA molecules Agarose concentration, the conformation of DNA, applied current
- Matrix of agarose gel works as a molecular filter through the DNA fragments can move upon the application of an electric current.
- A higher concentration of agarose results in stronger gels. i.e. spaces between the cross-linked molecules are less and therefore smaller DNA fragments can easily move through these gaps.
- When the size of DNA grows, it becomes more difficult for the DNA to travel through the spaces. Likewise, the lower concentration of agarose aids in the movement more large DNA pieces since the distance between the cross-linked molecules are larger.
- The progression of electrophoresis in gels is tracked by monitoring the movement of an invisible color (tracking the dye) across the gel.
- Two of the most commonly used dyes are Xylene Cyanol and Bromophenol blue, both of which travel at the same rate as double-stranded DNA with sizes of 5000 bp and 300 bp, respectively. The dyes used to track are negatively charged low molecular weight compounds that are added to each sample prior to the beginning of the run. Once the dye is able to reach the anode, the run ends.
- Because DNA isn’t naturally colored so it won’t be evident in the gel. Therefore, the gel, following electrophoresis, gets stained by an in-specific dye for the DNA.
- Distinction bands are visible in the presence of enough DNA that binds the dye to reveal it otherwise, the band cannot be observed.
- The gel is viewed against a white background, where DNA appears as dark-coloured bands.
- Alternately you can use an intercalating dye, such as Ethidium bromide can be added to the agarose gel. The the location of bands is identified by examining the gel in UV which is when DNA reflects.

current and can be visualized by using ethidium bromide which fluoresces under the ultraviolet
light | Image Source: https://himedialabs.com/TD/HTBM001.pdf
Take note: Ethidium bromide is to be handled with care since it can cause mutagenesis and is carcinogen. Use gloves when working with EtBr solutions and gels stained by EtBr.
Summery: DNA, RNA, and proteins are negatively charged molecules that can be segregated based on their size and charge through the application of an electric field. In agarose gel electrophoresis, the gel matrix is composed of agarose, a polysaccharide derived from seaweed that forms a porous structure when combined with water. The agarose gel functions as a sieve, permitting tiny molecules to pass through with greater ease than larger molecules. By placing the gel in a buffer solution and applying an electrical current across the gel using two electrodes, an electric field is created. The sample of DNA, RNA, or protein is combined with a loading buffer containing a tracking reagent that permits the electrophoresis to be monitored. After loading the sample onto the gel, the electric current is activated. Negatively charged sample molecules migrate through the gel to the positively charged electrode. The smaller molecules migrate through the gel more rapidly and reach the gel’s end first, while the larger molecules migrate more slowly and form distinct bands on the gel. A staining agent, such as ethidium bromide, which attaches to nucleic acid molecules and fluoresces under UV light can be used to visualise the bands.
Factors determine the rate of migration of DNA through agarose gels
The following elements influence the rate of transfer of DNA through agarose gels.
- Molecular size of DNA: Fragments of DNA that are linear move through agarose gels, with an amount of mobility that is related to their log10 molecular weight. Larger molecules travel slower because of the more frictional drag when they travel through the gel’s pores less efficiently than smaller molecules. Circular DNA forms migrate in agarose different from linear DNA with the same mass. Undigested plasmids travel faster that the identical plasmid if linearized.
- Agarose Concentration: Through the use of gels that contain different concentrations of agarose, DNA fragments that are various sizes can be separated. A higher concentration of agarose aids in the separation of small DNA and lower concentrations of agarose allow the resolution of large DNA.
- Electrophoresis buffers: Many buffers have been suggested for the electrophoresis of DNA. The most popular is The TAE (Tris-acetate-EDTA) as well as the TBE (Tris-borate-EDTA). DNA fragments move in different ways within the two buffers because of differences in the strength of ions. Buffers don’t just create the pH, they also supply Ions that help to improve conductivity.
- Effect of Ethidium bromide: it’s a is a fluorescent dye that emits light between 254-366nm. It is intercalated between the nucleic acids’ bases and permits the very efficient analysis of DNA fragments inside gels using an ultraviolet transilluminator. The binding of ethidium chloride to DNA alters its weight and rigidity and, consequently, its mobility.
- Voltage: The voltage applied to the gel increases, larger fragments move faster than smaller pieces. The most precise resolution of fragments greater than 2 kb is achieved by applying no more than 5 volts per cent on the gel (the cm value represents the distance between two electrodes, it isn’t the entire length).
Requirement for Gel electrophoresis
To conduct agarose gel electrophoresis, the following equipment and supplies are required:
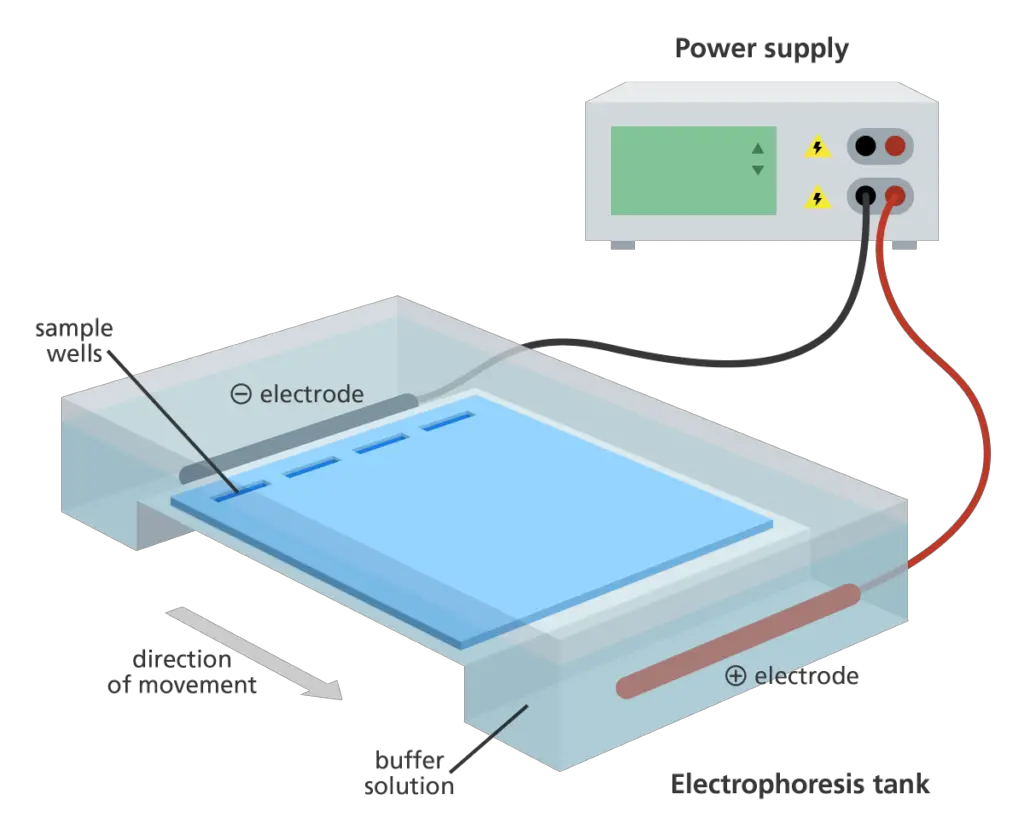
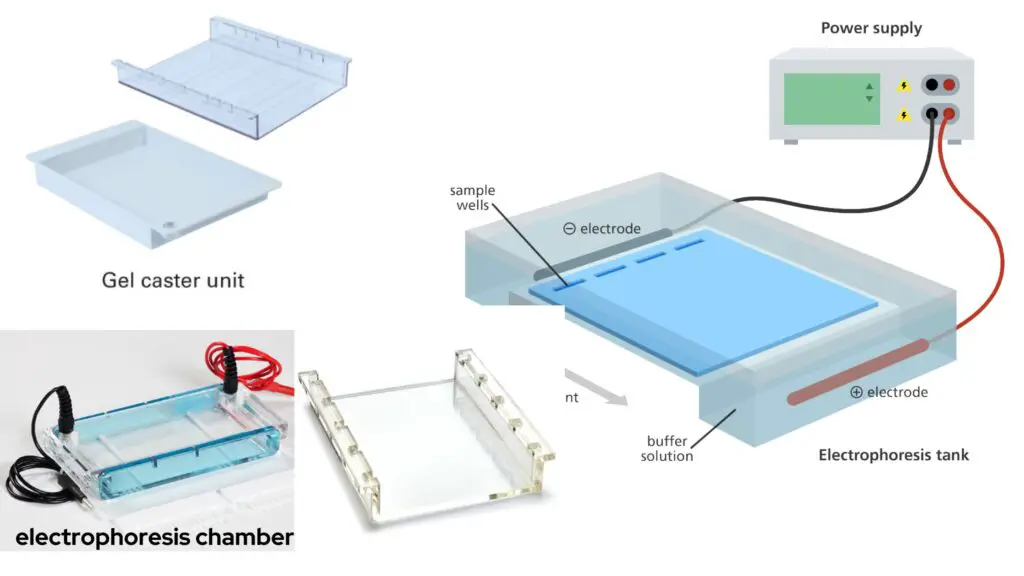
- Electrophoresis chamber and power supply: This device generates a potential difference across the gel, allowing the movement of charged molecules during electrophoresis.
- Gel casting trays: These trays are made of UV-transparent plastic and come in various sizes. They are used to hold the molten agarose gel during gel preparation. The open ends of the trays are sealed with tape while the gel is being cast and are removed prior to electrophoresis.
- Sample combs: These combs are placed in the gel casting trays before pouring the molten agarose. The combs create wells in the gel where the samples will be loaded.
- Electrophoresis buffer: The buffer is essential for creating the appropriate ionic environment for the movement of charged molecules through the gel. Commonly used buffers include Tris-acetate-EDTA (TAE) or Tris-borate-EDTA (TBE).
- Loading buffer: This buffer is mixed with the samples before loading them into the wells. It usually contains a dense substance like glycerol, which helps the samples sink into the wells. Additionally, one or two tracking dyes are included in the loading buffer to monitor the progress of electrophoresis.
- Staining: Staining is performed to visualize the DNA molecules within the gel. One commonly used stain is ethidium bromide (EtBr), which fluoresces under ultraviolet light when bound to DNA. Alternatively, DNA can be stained after electrophoretic separation by soaking the gel in a solution of EtBr. Another fluorochrome called 1-anilino 8-naphthalene sulphonate can also be used to detect DNA.
- Transilluminator: This is an ultraviolet light box used to visualize the stained DNA in the gel. When the gel is placed on the transilluminator and exposed to ultraviolet light, the DNA bands appear as fluorescent bands that can be observed and documented.
In addition to the specific equipment and supplies, other utilities such as pipettes, tips, flasks, and a weight balance may be necessary for sample preparation and handling.
The entire process of agarose gel electrophoresis typically takes around 70 to 90 minutes to complete.
Kit Components of Agarose gel electrophoresis
A standard agarose gel electrophoresis kit includes the following components:
- Agarose: Agarose is the major component of the gel matrix and is a polysaccharide obtained from seaweed. During electrophoresis, it generates a porous structure through which DNA molecules can travel.
- Agarose gel electrophoresis instrument: The electrophoresis chamber and power supply used to generate an electric field across the gel are referred to as the agarose gel electrophoresis apparatus. The gadget supplies the required voltage and current to allow charged molecules to flow through the gel.
- Agarose gel electrophoresis buffer: The kit comprises a particular buffer solution, such as Tris-acetate-EDTA (TAE) or Tris-borate-EDTA (TBE), which maintains the pH and supplies ions for conducting electricity during electrophoresis. The buffer is required to create the proper ionic environment for DNA molecule movement.
- DNA Gel loading dye: This dye is added to the DNA samples before they are loaded into the gel’s wells. Tracking dyes, such as bromophenol blue or xylene cyanol, are present in the loading dye and enable for visual monitoring of the electrophoresis progress. The colours travel at varying rates through the gel, demonstrating the mobility of DNA fragments.
- EtBr (Ethidium Bromide): EtBr is a fluorescent dye that is often used to stain DNA on agarose gels. It intercalates between DNA base pairs and fluoresces under UV light. The kit may include a small amount of EtBr or another DNA stain for observing the DNA bands after electrophoresis.
- Powerpack: Included in the kit is a powerpack, which is a device that controls and supplies the proper electrical current and voltage to the gel during electrophoresis. The powerpack maintains a continuous and controlled flow of energy across the gel for efficient DNA fragment separation.
These components are commonly included in an agarose gel electrophoresis kit, which provides researchers with the resources they need to perform DNA analysis and electrophoresis.
1. Agarose
Agarose is a polysaccharide generated from seaweed, specifically agarobiose, a polymer with a long chain. Agarose is made by removing agaropectin from agarose. This polysaccharide is made up of a linear chain of monomeric D-galactose and 3,6-anhydro-L-galactopyranose disaccharides.
When heated, agarose powder dissolves in water. It dissolves in a buffer solution when heated. When the dissolved agarose cools, it creates hydrogen bonds, which leads to cross-linking and polymerization. This cross-linking happens between adjacent agarose molecules, forming a three-dimensional matrix containing pores.
The size of these pores varies depending on the agarose content in the gel. Lower cross-linking results in bigger pore sizes, while higher cross-linking results in smaller pore sizes. These pores in the gel form routes via which DNA can travel.
The melting temperature of agarose is near to the boiling point, around 95oC. The gelling temperature, on the other hand, ranges between 37oC and 43oC. The pore size reduces as the agarose concentration increases. As demonstrated in the table below, different agarose concentrations are appropriate for various types of DNA samples:
- 0.8% agarose: Genomic DNA fragments larger than 1 kb.
- 1.0% agarose: PCR products and plasmid DNA ranging from 400 bp to 10 kb in 1.0% agarose.
- 2.0% agarose: PCR products ranging in size from 50 bp to 2 kb.
- 3.0% agarose: Restriction digestion results varied in fragment size from 10 to 1000 bp.
To achieve the best results, the concentration of the gel must be matched to the size of the DNA fragments. Satisfactory results cannot be obtained if the gel concentration does not correlate to the size of the DNA fragments. Larger DNA fragments, for example, cannot travel well in highly concentrated gels, resulting in a smudge of DNA rather than distinct bands.
Smaller DNA molecules travel faster than larger ones during electrophoresis. As a result, various DNA band patterns can be detected and analysed based on their diameters.
2. Agarose gel electrophoresis equipment
- The electrophoresis chamber, gel caster, gel comb, electrodes, and clamps are all part of the agarose gel electrophoresis equipment. These components are required for performing agarose gel electrophoresis.
- The electrophoresis chamber is usually made of high-quality acrylic material. It acts as a container for the agarose gel and a platform for electrophoresis of the gel. The chamber is intended to contain the gel and keep it steady during the procedure.
- The gel caster is used to prepare the gel. The gel caster is an important component that aids in the casting of the agarose gel. It serves as a frame or mould into which the liquid agarose solution is poured to create the gel. The gel caster keeps the gel in place and prevents it from leaking.
- To keep the gel tray in position, clamps are utilised in conjunction with the gel caster. These clamps assist in tightening the gel tray within the gel caster, guaranteeing a tight fit and preventing the gel from spilling or sliding during the casting process.
- The clamps are removed after the gel has formed and settled within the gel tray, and the gel tray is gently transported to the electrophoresis chamber. The chamber is filled with agarose gel electrophoresis buffer, which acts as a medium for the electric current to travel through during the electrophoresis process.
- To make wells or indentations in the agarose gel, gel combs are used. The DNA samples that will be analysed are placed into these wells. Before the liquid agarose solution solidifies, the gel combs are placed into it, creating slots or holes into which the DNA samples can be loaded.
- Two electrodes are installed in the electrophoresis chamber. A positive electrode is located at one end of the chamber, while a negative electrode is located at the other. These electrodes are linked to a power supply, which produces an electric field within the gel. Under the influence of the electric field, the DNA samples loaded in the gel’s wells will migrate through the gel, separating according to their sizes.
- Overall, the agarose gel electrophoresis equipment, which includes the electrophoresis chamber, gel caster, gel comb, electrodes, and clamps, is critical in the agarose gel electrophoresis process. It provides the tools and infrastructure required to accurately process and analyse DNA samples.
3. Agarose gel electrophoresis buffer
A essential component in DNA separation and analysis techniques is agarose gel electrophoresis buffer. It acts as a liquid medium that allows DNA to migrate across the gel matrix. In agarose gel electrophoresis, two buffer systems are often used: TAE (Tris-acetate EDTA) and TBE (Tris-borate EDTA).
One of the most important features of the electrophoresis buffer is its capacity to keep the medium’s pH stable. This is significant because DNA movement and separation are largely reliant on pH. The buffer aids in the creation of an appropriate environment for effective DNA mobility during electrophoresis.
The borate in TBE buffer might react with the glycerol in the DNA gel loading dye. This contact can interfere with the movement of DNA molecules, resulting in incorrect outcomes. As a result, TBE buffer is not usually the best choice for electrophoresis. TAE buffer, on the other hand, is often employed due to its compatibility with the gel loading dye and ability to deliver consistent and reproducible results.
The following ingredients are needed to make a 10X TAE buffer with a total capacity of 250ml:
- 400mM Tris-HCl
- Acetic acid (200mM)
- 10% EDTA
These ingredients should be combined, then distilled water (D/W) should be added to make a final volume of 250ml. To maintain consistent results, it is crucial to use only one type of buffer for both gel preparation and tank filling.
4. DNA gel loading dye
DNA gel loading dye is an essential component used in DNA analysis during gel electrophoresis. It contains two main constituents: Bromophenol blue and glycerol. The primary purpose of the loading dye is to monitor the migration of DNA through the gel while the electrophoresis process is taking place.
The inclusion of Bromophenol blue in the loading dye serves as a tracking dye. It runs ahead of the DNA fragments, allowing scientists to visualize the movement of the DNA during electrophoresis. By observing the migration of the tracking dye, researchers can determine the distance covered by the DNA in the gel, aiding in the analysis and interpretation of the results.
Another crucial component of the DNA gel loading dye is glycerol. Glycerol provides density to the DNA sample, enabling it to settle at the bottom of the well during loading. This ensures that the DNA stays localized within the gel well and facilitates efficient loading onto the gel. The glycerol also helps to increase the viscosity of the DNA sample, making it easier to handle and reducing the likelihood of sample loss or dispersion during the loading process.
The composition of a typical 1X loading dye solution includes:
- 0.042% (W/V) Bromophenol blue powder: This concentration of Bromophenol blue is sufficient to provide a visible dye front during electrophoresis.
- 2.5% Ficoll: Ficoll is a high-molecular-weight polymer that adds viscosity to the loading dye, aiding in the sample loading process.
- 0.042% (W/V) Xylene cyanol FF (optional): Xylene cyanol FF is an alternative tracking dye that can be added to the loading dye mixture. Its presence allows for visual confirmation of the dye migration.
- Final makeup with distilled water (D/W): The loading dye components are mixed together, and distilled water is added to reach the desired final volume.
5. EtBr
Ethidium bromide (EtBr) is a commonly used fluorescent dye in molecular biology and microbiology for visualizing and tagging DNA. One of the unique properties of EtBr is its ability to intercalate or insert itself between the DNA bases. This intercalation results in the dye becoming incorporated within the DNA molecule, thereby altering its optical properties. Specifically, when exposed to ultraviolet (UV) light, the EtBr-DNA complex emits a characteristic fluorescent color, allowing for easy detection and visualization of DNA bands in electrophoresis gels.
It is important to note that working with Ethidium bromide requires caution and adherence to safety protocols. Ethidium bromide is classified as a mutagenic compound, meaning it has the potential to cause changes in the genetic material, including DNA. To ensure personal safety, it is recommended to take appropriate precautions when handling EtBr.
Firstly, it is crucial to wear protective gear, including gloves, a mouth cap, and UV protective glasses, to minimize the risk of exposure. Gloves provide a barrier between the skin and the dye, preventing direct contact. A mouth cap helps to avoid accidental ingestion or inhalation of the dye, while UV protective glasses shield the eyes from harmful UV radiation.
Additionally, it is advisable to handle EtBr in a well-ventilated area or under a fume hood to minimize the inhalation of any potential fumes that may be released during handling.
Proper waste management is essential when working with EtBr. Dispose of any waste containing EtBr according to local regulations and guidelines for hazardous materials.
Although EtBr is mutagenic and requires caution, when handled appropriately, it remains a valuable tool in molecular biology and microbiology research for visualizing and analyzing DNA samples. Researchers and laboratory personnel should follow established safety protocols and take necessary precautions to ensure their safety while working with EtBr.

6. Powerpack
The power pack is an essential component in DNA electrophoresis, providing the necessary electric current to facilitate the separation and migration of DNA fragments through the agarose gel. It serves as the power source for the electrophoresis process, enabling the movement of charged DNA molecules in response to an electric field.
The power pack consists of two electrodes, typically referred to as the positive electrode (anode) and the negative electrode (cathode). These electrodes are connected to the respective terminals of the power pack. The anode is where the positively charged ions accumulate, while the cathode attracts the negatively charged ions.
When DNA samples are loaded into the wells of the gel, the power pack is activated, and an electric current is applied across the gel. The electric field generated between the electrodes creates a voltage gradient within the gel matrix. This voltage gradient causes the DNA fragments to migrate towards the opposite electrode based on their charge, size, and conformation.
The choice of voltage applied to the gel depends on the specific requirements of the DNA sample being analyzed. Common voltage options are 50V, 100V, or 120V. Lower voltages are often used for the separation of larger DNA fragments, while higher voltages can facilitate the separation of smaller DNA fragments. The voltage selection should be based on the desired resolution and separation efficiency of the DNA fragments.
It is important to note that while the power pack provides the electric current for DNA electrophoresis, other factors such as gel concentration, buffer composition, and run duration also impact the separation and resolution of DNA fragments. Optimal conditions for electrophoresis, including voltage, must be determined based on the specific experimental requirements and the DNA samples being analyzed.
Agarose Gel Electrophoresis Protocols
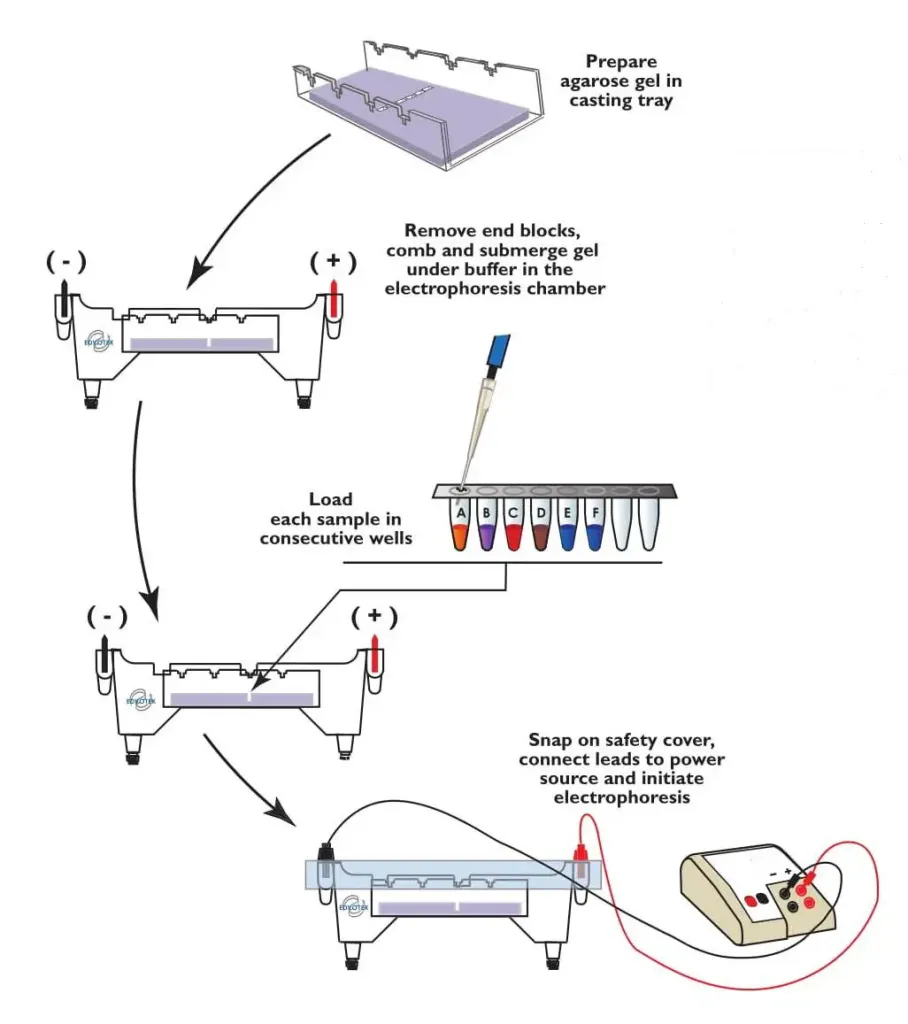
Preparation of 1% Agarose Gel
- Make 1X TAE preparation by diluting a suitable amount of buffer 50X. (For an experiment of one, about 200 ml of 1XTA is needed. Create 4ml of 50X TAE up to 200 ml using distilled water).
- Measure 0.5 grams of agarose and add it to 50ml of TAE 1X. This will yield the agarose gel at 1. Cook until the agarose gel dissolves completely and clear solutions are formed.
- Then, place the combs of electrophoresis in a position that is 2cm to the anode.
- Pour the solution of agarose into the tank’s central area at the point that it reaches 60 degrees Celsius.
- Do not create air bubbles.
- Its thickness must be between 0.5 up to 0.9 cm.
- Make sure the gel is kept at room temperature to allow the agarose to set.
- Pour 1X TAE into the gel tank until the buffer level is 0.5 to 0.8 centimeters above the gel’s surface.
- Carefully lift the combs up and ensure that the wells are unbroken.

Electrophoresis
- The power cable should be connected to an electrophoretic power source in accordance with the convention red anode, and black cathode.
- The samples should be loaded into those wells, in correct order.
- Set the voltage at 50 V and then turn on the power source.
- Turn off the power once the dye that tracks (bromophenol blue) in the well is 3/4th of the gel. It takes about one hour.
Staining and Visualization
- Prepare 1X staining dyes through diluting the 6X dye (1:6) with distillate water. (Approximately 50 milliliters of 1X staining dye is required for a single experiment. So, you should mix 8 ml of dye 6X up to 48 ml using distilled water).
- Make sure to transfer the gel (from the gel tank) into a tray that contains 1X staining solutions. Be sure that the gel is fully submerged.
- To achieve uniform staining, put the dish on a stand for around an hour, or shake it intermittently between 10 and 15 minutes.
- Dissolve the staining dye into the container. (The dye can be used two times). Clean the gel with tap water many times until the DNA appears as a dark spot against an unlit blue background.
Notably, Ethidium bromide could be used to visualize DNA fragments. Add Ethidium bromide to molten agarose until an amount of 0.5 Ug/ml (from an initial stock of 10 mg/ml of water) at temperatures approximately 50degC. Mix the gel and then cast it. The DNA sample are visible under UV light. They appear to be fluorescent. Destaining is not required in this situation.
Observation and Result of Agarose Gel Electrophoresis
The light reflection of stained gels with UV light (254-366 nm) permits DNA banding to be seen against a background that is free of dye. The gel image is recorded by making an Polaroid(tm) image or by using the gel documentation system.
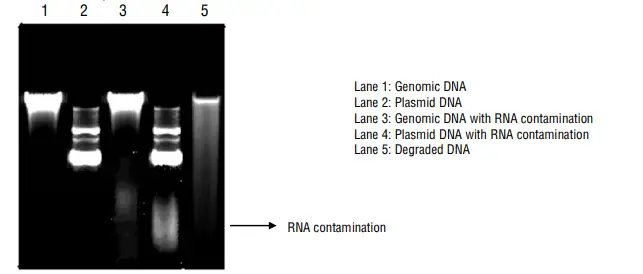
DNA concentrations can be estimated by
A. Taking absorbance at 260 nm.
1 absorbance (A) unit corresponds to the following concentration at 260 nm:
- 50 μg/ml for dsDNA
- 40 μg/ml for RNA
- 33 μg/ml for ssDNA
- 20-30 µg/ml for oligonucleotides
Although this technique is quick, nondestructive, and provides information about the sample’s purity (e.g., the presence of protein or organic contaminants), only estimates with concentrations of at least 1 g/ml are reliable. Moreover, this procedure cannot differentiate between DNA and RNA.
B. Intensity of Ethidium Bromide Fluorescence
The intensity of ethidium bromide fluorescence can be used to estimate the quantity of DNA in a sample (fluorescence emitted by ethidium bromide is proportional to the amount of DNA). The DNA concentration in a “unknown” solution can be estimated by comparing its fluorescence intensity to that of known quantities of DNA of comparable size. If a DNA sample is contaminated with other compounds that absorb in the UV range or is too dilute to measure at 260 nm, this method is useful.
Types of electrophoresis
There are several types of electrophoresis techniques commonly used in biological and biochemical research. Here are some of the most widely employed types:
- Agarose Gel Electrophoresis: As described earlier, agarose gel electrophoresis is used to separate DNA fragments based on their size. It is commonly used for DNA analysis, such as genotyping, DNA sequencing, and restriction enzyme digestion.
- Polyacrylamide Gel Electrophoresis (PAGE): PAGE is a type of gel electrophoresis that uses a polyacrylamide gel instead of agarose. It provides higher resolution and is primarily used for separating proteins, RNA, and smaller DNA fragments. It is commonly used in protein analysis, such as SDS-PAGE (sodium dodecyl sulfate-polyacrylamide gel electrophoresis) for protein size estimation and protein identification.
- Capillary Electrophoresis (CE): Capillary electrophoresis utilizes a narrow capillary tube filled with a separation buffer as the separation medium. It allows for highly efficient separation of molecules based on charge-to-mass ratio. CE is used for DNA sequencing, fragment analysis, protein analysis, and the separation of various small molecules, such as ions, carbohydrates, and pharmaceutical compounds.
- Isoelectric Focusing (IEF): IEF separates molecules based on their isoelectric points (pI), which is the pH at which they have no net charge. In IEF, a pH gradient is established in a gel or capillary, and charged molecules migrate towards their respective pI positions, resulting in their separation. IEF is commonly used for the separation of proteins and peptides based on their charge differences.
- Two-Dimensional Electrophoresis (2DE): This technique combines two different separation methods to achieve higher resolution. Typically, 2DE involves separating proteins by isoelectric focusing in the first dimension, followed by SDS-PAGE in the second dimension. It allows for the separation of complex protein mixtures and is commonly used in proteomics research.
- Immunoelectrophoresis: This technique combines electrophoresis with immunodiffusion to separate and identify proteins based on their antigen-antibody interactions. It is used for the analysis of proteins, particularly for detecting and quantifying specific antigens or antibodies in biological samples.
Role Of EtBr In Agarose Gel Electrophoresis
EtBr (ethidium bromide) is commonly used in agarose gel electrophoresis to visualize DNA. It is an aromatic molecule with a heterocyclic moiety. EtBr has the ability to intercalate or insert itself between the base pairs of DNA molecules, resulting in fluorescence when exposed to ultraviolet (UV) light. This property makes it a valuable tool in molecular biology techniques, including gel electrophoresis.
In gel electrophoresis, a small amount of EtBr is added to the agarose gel or to the DNA sample before loading it onto the gel. During electrophoresis, an electric current is applied, causing the DNA fragments to migrate through the gel matrix based on their size. As the DNA fragments move through the gel, they are exposed to the EtBr, which intercalates between the base pairs and binds to the DNA molecules.
After electrophoresis, the gel is visualized under UV light. The EtBr-DNA complexes emit orange fluorescence, allowing the DNA bands to be observed and photographed. By comparing the position and intensity of the DNA bands, valuable information about the size and quantity of DNA fragments can be obtained.
EtBr is also used in the karyotyping process of cytogenetics, which involves the analysis of chromosomes. It helps in visualizing the DNA present in the chromosomes and assists in identifying structural abnormalities or chromosomal rearrangements.
It is worth noting that EtBr has been associated with potential health hazards due to its mutagenic and carcinogenic properties. Therefore, precautions should be taken when handling and disposing of EtBr and appropriate safety measures should be followed to minimize exposure.
In summary, EtBr is an essential component in agarose gel electrophoresis for visualizing DNA fragments. Its ability to intercalate with DNA and emit fluorescence under UV light makes it a valuable tool in molecular biology and cytogenetics for studying DNA and chromosomes.
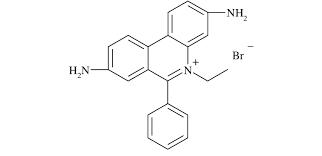
- Chemical name: Ethidium bromide
- Abbreviation: EtBr, etbr, EB
- Chemical formula: C21H20N3Br
- Molecular weight: 394.4
- Color emission: orange
Role of EtBr in gel electrophoresis
- EtBr, short for ethidium bromide, plays a crucial role in gel electrophoresis as a color agent and separating agent. Gel electrophoresis is a technique used to separate biomolecules based on their charges and sizes. DNA fragments, for example, migrate through a gel matrix towards the positive charge under the influence of an electric current.
- One limitation of DNA is that it lacks color, making it difficult to visualize with the naked eye. This is where EtBr comes in. EtBr is a fluorescent dye that intercalates between DNA base pairs and emits fluorescence when exposed to ultraviolet (UV) light. It imparts an orange color to the DNA, which can be observed using a standard orange filter. The fluorescence emitted by EtBr occurs at a wavelength of 470nm.
- The structure of EtBr consists of a tricyclic phenanthridine ring system. This ring system interacts with the hydrophobic interior of DNA bases through strong van der Waals bonds. Approximately one EtBr molecule binds to every 2.5 base pairs of DNA. The hydrophobic environment present between the base pairs enhances the intensity of fluorescence. Water, on the other hand, acts as an efficient quencher of fluorescence. When EtBr intercalates between the base pairs, it removes water and emits fluorescence more intensely.
- The intensity of fluorescence increases with the concentration of EtBr. The standard concentration of EtBr used in gel electrophoresis is typically 0.5μg/ml for a 50ml gel. EtBr enables the visualization and identification of various DNA molecules, such as PCR products, genomic DNA, restriction digestion products, and RNA.
- However, it is important to note that EtBr can alter DNA properties, including mobility, molecular weight, and charge. A study conducted by Sigmon J and Larcom LL in 1966 investigated the effect of EtBr intercalation on double-stranded DNA mobility. They examined various concentrations of EtBr and found that as the concentration increases, the mobility of DNA decreases. Despite the observed alterations in DNA properties, EtBr is still widely used for DNA confirmation studies.
- There are two common methods of using EtBr in gel electrophoresis. The first method involves directly adding EtBr into the gel. A predetermined amount of agarose is dissolved in a buffer and heated until a clear solution is obtained. Once the temperature becomes manageable, EtBr is added to the solution and mixed well through gentle shaking. This method is widely accepted in molecular laboratories. However, it has some limitations, including decreased EtBr activity at higher temperatures and poor spreading if the solution cools rapidly.
- The second method is gel staining by EtBr. After the gel electrophoresis run is complete, the gel is stained overnight with a mild EtBr solution. This method is time-consuming and not recommended due to the carcinogenic nature of EtBr.
- In conclusion, EtBr serves as both a color agent and a separating agent in gel electrophoresis. It enables the visualization and identification of DNA fragments through its fluorescence under UV light. Despite its ability to alter DNA properties, EtBr remains widely used in DNA confirmation studies. However, precautions should be taken due to its carcinogenic nature.
Role of EtBr in cytogenetic
- EtBr, also known as ethidium bromide, plays a significant role in cytogenetics, particularly in high-resolution banding techniques used to study chromosome structure and detect chromosomal abnormalities. In conventional cytogenetics, metaphase chromosomes are cultured and fixed to analyze abnormalities associated with various syndromes.
- Metaphase chromosomes are highly condensed, making it difficult to interpret minor variations. EtBr can be used to release chromosomal condensation and elongate the chromosomes, making them more visible and suitable for screening minor variations. This method, known as high-resolution banding, allows for the detection of smaller deletions, duplications, and insertions.
- To increase the number of chromosomal bands and enhance the resolution of the results, EtBr is added before harvesting the chromosomes. Typically, the chromosomes are cultured for 72 hours, and EtBr is added at around the 70th hour of harvesting. Once added, EtBr intercalates between DNA bases, preventing DNA folding and condensing. The resulting loosely packed DNA leads to longer chromosomes and increased band sizes. The greater the number of chromosome bands, the higher the accuracy of the results.
- While there is some debate among scientists regarding the specific role of EtBr in chromosomal elongation, a study conducted by Shinichi Misawa et al. in 1986 provided evidence supporting its importance. The researchers treated cultured human bone marrow cells with different doses of EtBr two hours before harvesting. They found that the addition of EtBr significantly increased the yield of results compared to cultures without EtBr. With the addition of 10 μg/ml of EtBr, they achieved over 400 bands per haploid cell, representing a 2.9-fold increase in yield.
- EtBr treatment was found to decrease chromosome condensation and increase chromosome length, leading to improved quality of chromosomes in high-resolution banding. This finding highlights the value of EtBr in cytogenetic studies and its role in enhancing the resolution and accuracy of chromosomal analysis.
Limitations of EtBr
- EtBr is a fluorescent dye with a diminished half-life.
- Mutagenic, because it can bind to DNA.
- EtBr is both carcinogenic and teratogenic. However, the carcinogenicity of EtBr has not been demonstrated. It affects through inhalation, skin absorption, and ingestion.
- It cannot be used for extended periods of time because its activity decreases over time.
DNA Gel Loading Dye – Bromophenol Blue And Xylene Cyanol
DNA gel loading dye is a crucial component in agarose gel electrophoresis, used to monitor the migration of DNA. Two commonly used dyes in the loading dye are bromophenol blue and xylene cyanol. Let’s delve into their roles and properties:
- Bromophenol Blue: Bromophenol blue is a pH indicator that is widely utilized to visualize DNA during agarose gel electrophoresis. It appears as light pink to purple crystals and is soluble in water. This dye has several applications beyond gel electrophoresis, including as a color indicator, an acid-base pH indicator, and a biological stain.
Bromophenol blue exhibits a yellow color at a pH of around 3 and transitions to a blue color at pH values above 4.8. In gel electrophoresis, it serves as a tracking dye, allowing researchers to monitor the progress of DNA migration within the gel. Its movement through the gel helps to estimate the migration distance of DNA fragments and assess the progress of the electrophoresis run.
- Xylene Cyanol: Xylene cyanol is another commonly used dye in DNA gel loading dye. Like bromophenol blue, it is a tracking dye that aids in visualizing the migration of DNA fragments during electrophoresis. Xylene cyanol is a blue or dark green powder and is soluble in water.
In agarose gel electrophoresis, xylene cyanol migrates at a rate that is generally slower than bromophenol blue, allowing for better differentiation and estimation of DNA migration distances. It serves as an additional reference point and aids in monitoring the progress of DNA migration within the gel.
Both bromophenol blue and xylene cyanol are incorporated into the loading dye in appropriate concentrations to provide visible reference points during gel electrophoresis. Their distinct colors and migration patterns aid in visualizing the movement of DNA fragments, enabling researchers to assess the success of the electrophoresis run and analyze the size of DNA fragments.
It is worth noting that the specific concentrations and ratios of bromophenol blue and xylene cyanol in the loading dye can vary depending on the specific protocol or laboratory preferences. However, their fundamental role remains consistent as tracking dyes in gel electrophoresis, facilitating the analysis and interpretation of DNA migration.

Function of gel loading dye
Gel loading dye plays a crucial role in agarose gel electrophoresis by serving three important functions:
- Color Indicator: DNA is colorless and odorless, making it difficult to visualize its migration in a gel. Gel loading dye contains specific chemicals, such as bromophenol blue (BPB), that migrate ahead of the DNA fragments during electrophoresis. BPB acts as a color indicator, allowing researchers to monitor the progression of electrophoresis. Its migration can be observed, indicating that the DNA is also migrating through the gel.
- Density Agent: DNA is less dense than the running buffer used in electrophoresis, which can cause it to diffuse and disperse in the buffer. The loading dye contains substances like Ficoll or glycerol that provide density to the DNA sample. This helps the DNA settle at the bottom of the well and prevents it from diffusing into the buffer. By settling the DNA, the loading dye ensures that the DNA migrates properly and produces clear and well-defined bands on the gel.
- Detection of Overflow, Cross-Contamination, or Leakage: The color composition of the loading dye aids in detecting any issues related to sample overflow, cross-contamination, or leakage. If there is an unintended release or mixing of samples, the distinct color of the loading dye can help identify and monitor such occurrences.
The composition of the loading dye is crucial to achieve clear and accurate results. Maintaining the proper concentration of the dye components is essential for obtaining optimal clarity and visibility of DNA migration bands on the gel.
In summary, gel loading dye acts as a color indicator, provides density to the DNA sample, and helps detect any sample overflow or cross-contamination. By fulfilling these functions, it facilitates the monitoring and analysis of DNA migration during agarose gel electrophoresis.
DNA gel loading dye composition
The composition of a typical 1X DNA gel loading dye includes:
- 0.042% (W/V) Bromophenol blue powder: Bromophenol blue is a commonly used dye in loading dye. It imparts a distinct color and migrates ahead of DNA fragments during electrophoresis, allowing for easy visualization of the DNA migration.
- 2.5% Ficoll: Ficoll 400, a hydrophilic, high molecular weight polysaccharide, is included in the loading dye to provide density to the DNA sample. It helps the DNA settle at the bottom of the well and prevents diffusion into the buffer.
- 0.042% (W/V) Xylene cyanol FF (optional): Xylene cyanol is an additional dye that can be added to the loading dye, although it is not always included. Like bromophenol blue, xylene cyanol helps monitor the progression of electrophoresis and can migrate ahead of DNA fragments.
- Final makeup with distilled water (D/W): The loading dye is made up to the final volume using distilled water. This ensures the proper concentration and compatibility of the dye components.
To make an effective DNA gel loading dye, certain characteristics are desired:
- The combination of bromophenol blue and xylene cyanol enhances the effectiveness and quality of the results. These dyes should migrate ahead of the DNA fragments for proper visualization under UV light.
- The loading dye should carry a negative charge, primarily through bromophenol blue, allowing it to migrate toward the positive node in agarose gel electrophoresis.
- The components of the loading dye should not interact with DNA or agarose, as this can affect the DNA’s structure, mobility, and the gel’s clarity.
- Temperature stability is essential to ensure that the loading dye remains stable even at higher voltages or increased temperatures during electrophoresis.
Additional components like EDTA and Tris may be added to the loading dye recipe, although they are optional. EDTA helps inhibit nuclease activity, while Tris maintains the pH of the loading dye. However, since EDTA and Tris are already present in the electrophoresis buffer, their inclusion in the loading dye can be omitted.
Ficoll 400 is highly recommended as a polysaccharide in the loading dye due to its hydrophilic nature, higher molecular weight, and non-reactivity with DNA, buffer, or agarose. This makes Ficoll 400 an ideal choice for providing density to the DNA sample.
In summary, the composition of DNA gel loading dye typically includes bromophenol blue, Ficoll 400, and optionally xylene cyanol. The dye should possess the desired characteristics of migrating ahead of DNA, carrying a negative charge, not interacting with DNA or agarose, and being temperature-stable for optimal results in gel electrophoresis.
Agarose Gel Electrophoresis Buffer
Agarose gel electrophoresis requires the use of a specific buffer to create the optimal environment for the separation of DNA, RNA, or protein molecules. The two commonly used buffer solutions for this purpose are Tris-borate EDTA (TBE) and Tris-acetate EDTA (TAE).
TBE buffer is composed of Tris base, boric acid, and EDTA. Tris base acts as a weak base to maintain the pH of the buffer solution, while boric acid provides buffering capacity. The addition of EDTA, a chelating agent, helps protect the DNA from degradation by binding to divalent metal ions that could catalyze nucleic acid hydrolysis. TBE buffer is particularly well-suited for high-resolution DNA separations due to its buffering capacity and ability to maintain stable pH conditions.
On the other hand, TAE buffer consists of Tris base, glacial acetic acid, and EDTA. Tris base functions as a weak base to maintain pH stability, while glacial acetic acid provides buffering capacity. TAE buffer is commonly used for routine DNA electrophoresis and is less prone to precipitate compared to TBE buffer.
To prepare a 10X TBE or TAE buffer, the appropriate amounts of Tris base, boric acid (for TBE), glacial acetic acid (for TAE), and EDTA are dissolved in distilled water. The pH of the buffer solution is then adjusted using concentrated acids or bases to achieve the desired pH level. Finally, the volume is brought up to the desired final volume with distilled water.
When preparing the working buffer solution for agarose gel electrophoresis, the 10X concentrated TBE or TAE buffer is typically diluted to a 1X concentration. The 1X buffer is used to create the electrophoresis chamber and is mixed with the agarose gel to ensure proper conductivity and pH stability during the separation process.
The choice between TBE and TAE buffer depends on various factors, including the desired resolution, compatibility with downstream applications, and personal preference. Both buffer systems effectively support the migration of DNA molecules through the agarose gel matrix and enable the separation and analysis of nucleic acids.
In summary, the agarose gel electrophoresis buffer, whether TBE or TAE, plays a critical role in creating the optimal conditions for the separation of DNA, RNA, or protein molecules. These buffer systems provide pH stability, buffering capacity, and protection against DNA degradation, ensuring accurate and reliable results in the electrophoresis process.
Importance of gel electrophoresis buffer
The gel electrophoresis buffer plays a crucial role in maintaining the optimal conditions for the separation and analysis of DNA, RNA, or protein molecules. Its importance can be understood through the following points:
- pH Maintenance: The buffer solution helps maintain a nearly neutral pH throughout the electrophoresis process. The presence of a weak acid and base in the buffer helps in buffering the system and prevents significant fluctuations in pH. A stable pH is essential for maintaining the net charge of the molecules being separated, which in turn ensures their proper migration and separation within the gel.
- Net Charge Control: The pH stability provided by the buffer solution helps regulate the net charge of the molecules being analyzed. The net charge of DNA, RNA, or protein molecules depends on the pH of the surrounding medium. By maintaining a neutral pH, the buffer ensures that the molecules retain their expected charge, facilitating their migration towards the appropriate electrode during electrophoresis.
- Protection against Enzymatic Degradation: The DNA gel electrophoresis buffer serves as a protective medium that prevents DNA molecules from being degraded by DNases. DNases are enzymes that can break down DNA, leading to the loss of integrity and accurate analysis of the samples. The buffer provides a constant liquid environment that helps protect the DNA from enzymatic attacks, maintaining the integrity of the molecules throughout the electrophoresis process.
- Temperature Control: During electrophoresis, higher voltages can generate heat, which may potentially denature DNA molecules. The buffer acts as a temperature control medium, helping to dissipate heat and regulate the temperature within the gel. This prevents excessive heating and ensures that the DNA molecules remain in their native state, allowing for accurate analysis.
- Charge Distribution: The buffer solution plays a crucial role in distributing the electrical charge evenly across the gel during electrophoresis. This even distribution of charge helps in achieving consistent migration rates for the molecules being separated and ensures the production of clear and well-defined bands on the gel.
In summary, the gel electrophoresis buffer is vital for maintaining a stable pH, controlling the net charge of the molecules, protecting against enzymatic degradation, regulating temperature, and ensuring even charge distribution. These functions collectively contribute to the success and accuracy of the electrophoresis process, allowing for reliable separation, analysis, and characterization of DNA, RNA, or protein samples.
Role of TBE/ TAE buffer in agarose gel electrophoresis
The TBE (Tris-Borate-EDTA) and TAE (Tris-Acetate-EDTA) buffers are commonly used in agarose gel electrophoresis to provide the necessary pH stability and buffering capacity. The role of these buffers can be understood as follows:
- pH Maintenance: Both TBE and TAE buffers contain Tris, which acts as a strong base, and borate or acetate, which act as acids. The combination of these components helps maintain the pH of the buffer in the neutral range of 8 to 8.5. This slightly alkaline pH is optimal for DNA stability and proper separation during electrophoresis.
- DNA Protection: The addition of EDTA in the buffer serves an important role. EDTA is a chelating agent that binds to divalent cations, such as Mg2+, which are required as cofactors for enzymes like DNases. By chelating the Mg2+ ions, EDTA inhibits the activity of DNases and protects the DNA from enzymatic degradation during the electrophoresis process.
- Charge Neutralization: During electrophoresis, water molecules dissociate into H+ and OH- ions under the influence of the electrical current. The H+ ions can react with the negatively charged phosphate groups (PO3-) of DNA, potentially affecting its integrity and migration. The negatively charged buffer components in TBE or TAE neutralize the charge of water molecules, preventing them from interacting with DNA and ensuring its protection during electrophoresis.
It is worth noting that TAE and TBE buffers have some differences that researchers consider when selecting the appropriate buffer for their specific needs.
- TAE buffer is known to interfere with certain enzymatic reactions due to the presence of acetate ions. However, it provides effective protection for DNA during electrophoresis.
- TBE buffer, on the other hand, has a higher buffering capacity due to the presence of borate ions. However, borate can react with the sugar backbone of DNA and affect its integrity. Additionally, if glycerol is a component of the DNA gel loading dye, borate can react with it and decrease the activity of the loading dye.
Considering these factors, if glycerol is the primary component in the DNA gel loading dye, TAE buffer is generally recommended to ensure optimal results. However, if enzymatic reactions need to be carried out after electrophoresis or if a higher buffering capacity is desired, TBE buffer can be used with caution.
Overall, the choice between TAE and TBE buffers depends on the specific experimental requirements, taking into account factors such as DNA protection, compatibility with enzymatic reactions, buffering capacity, and the presence of other components in the experimental setup.
Properties of TAE buffer
TAE buffer (Tris-Acetate-EDTA) is a commonly used buffer system in agarose gel electrophoresis for DNA separation. The characteristics and benefits of TAE buffer can be summarized as follows:
- Lower Buffering Capacity: TAE buffer has a lower buffering capacity compared to TBE buffer. This means that its ability to maintain a stable pH over repeated use is limited. With prolonged use or in large-scale electrophoresis, the buffering capacity of TAE buffer can become exhausted, leading to pH fluctuations and potential DNA damage. Therefore, TAE buffer is typically recommended for smaller-scale experiments or single-use applications.
- Higher Conductivity: TAE buffer exhibits higher conductivity compared to TBE buffer. The higher conductivity allows for faster migration of double-stranded DNA (dsDNA) molecules through the gel matrix during electrophoresis. This makes TAE buffer particularly suitable for applications where faster separations are desired.
- Enhanced DNA Recovery: TAE buffer facilitates the easy recovery of DNA from the gel after electrophoresis. The low viscosity of TAE buffer and its compatibility with various DNA extraction methods make DNA retrieval more efficient. This is especially advantageous when downstream applications, such as DNA purification or enzymatic reactions, are planned following gel electrophoresis.
- Cost-Effectiveness: TAE buffer is considered a cost-effective option in comparison to other buffer systems. It typically requires fewer components and has a lower overall cost. Additionally, the working solution requirement for TAE buffer is lower, as a 0.5X concentration of the buffer is often sufficient for optimal performance. This makes TAE buffer an economical choice for routine agarose gel electrophoresis experiments.
Overall, TAE buffer offers advantages such as faster DNA migration, easier DNA recovery, and cost-effectiveness. However, researchers should be aware of its lower buffering capacity and potential limitations in larger-scale or long-duration experiments. By considering these factors, scientists can make an informed choice regarding the use of TAE buffer for their specific gel electrophoresis applications.
Properties of TBE buffer
TBE buffer (Tris-Borate-EDTA) is another commonly used buffer system in agarose gel electrophoresis for DNA separation. Let’s explore the characteristics and considerations associated with TBE buffer:
- Higher Buffering Capacity: TBE buffer has a higher buffering capacity compared to TAE buffer, primarily due to the presence of borate ions. The higher buffering capacity allows TBE buffer to maintain a stable pH over a longer period, even with repeated use. This makes TBE buffer well-suited for larger-scale experiments or prolonged electrophoresis runs.
- Lower Conductivity: TBE buffer exhibits lower conductivity compared to TAE buffer. This means that the migration of double-stranded DNA (dsDNA) through the gel matrix is slower in TBE buffer. Consequently, TBE buffer is often preferred for applications that require better resolution of longer DNA fragments, as the slower migration allows for clearer separation of DNA bands.
- Enhanced DNA Integrity: The presence of borate ions in TBE buffer can inhibit certain DNA enzymes, which can help preserve the integrity of DNA during electrophoresis. This is particularly beneficial when working with delicate or sensitive DNA samples that may be susceptible to enzymatic degradation. The use of TBE buffer can contribute to maintaining the quality and integrity of DNA molecules.
- Cost and Working Solution Requirement: TBE buffer tends to be slightly more expensive than TAE buffer, primarily due to the cost of borate. Additionally, TBE buffer is typically used at a 1X concentration, which requires a higher volume of the buffer for optimal performance. This means that the working solution requirement for TBE buffer is higher compared to TAE buffer, which may have cost implications in large-scale experiments.
- Recovery Rate: Borate ions present in TBE buffer can interact with DNA molecules, making the recovery of DNA from the gel less efficient compared to TAE buffer. This can pose challenges when downstream applications require the extraction or purification of DNA fragments from the gel.
In summary, TBE buffer offers a higher buffering capacity, better resolution for longer DNA fragments, and potential advantages for preserving DNA integrity. However, it also has lower conductivity, higher cost, higher working solution requirement, and a lower DNA recovery rate. Researchers should consider the specific requirements of their experiments and choose the buffer system that best suits their needs. While TAE buffer is generally recommended due to its cost-effectiveness and ease of use, TBE buffer may be preferred when high-resolution separation of longer DNA fragments is crucial.
Preparation of TAE/ TBE buffer
The preparation of TAE (Tris-Acetate-EDTA) or TBE (Tris-Borate-EDTA) buffer is crucial for maintaining the appropriate pH and ionic conditions during agarose gel electrophoresis. Here’s a step-by-step guide for preparing 10X TAE/TBE buffer:
Preparation of 10X TBE buffer (for 250ml):
- Measure the following quantities of reagents:
- Tris: 900mM
- Boric Acid: 890mM
- EDTA: 2mM
- Add distilled water to make the final volume of 250ml.
- Combine Tris, boric acid, and EDTA in a container and mix until the EDTA is completely dissolved in water.
- Adjust the pH of the solution to approximately 8.0 to 8.5 using a pH meter or pH indicator strips.
- Once the 10X TBE buffer is prepared, take 10ml of the stock solution and dilute it with 90ml of distilled water to obtain 100ml of 1X TBE buffer.
Note: For larger volumes, the same ratio can be used. For example, for 1000ml of TBE buffer, mix 100ml of 10X TBE buffer with 900ml of distilled water.
Preparation of 10X TAE buffer (for 250ml):
- Measure the following quantities of reagents:
- Tris: 400mM
- Acetic Acid: 200mM
- EDTA: 10mM
- Add distilled water to make the final volume of 250ml.
- Combine Tris, acetic acid, and EDTA in a container and mix thoroughly until all components are dissolved.
- The preparation of 1X TAE buffer is similar to that of TBE buffer. Take 10ml of the 10X TAE buffer stock solution and dilute it with 90ml of distilled water to obtain 100ml of 1X TAE buffer.
It is important to note that freshly prepared buffers yield better results, and for optimal performance, it is recommended to prepare a fresh 1X buffer for each run. The buffering capacity of the buffer may decrease over time, affecting the quality of DNA bands observed. If reusing the buffer, it is advisable to do so for standardization and confirmation studies rather than critical or sensitive experiments.
Additionally, it is crucial to accurately weigh each chemical used in the buffer preparation, as accurate weighing ensures reliable and reproducible results. Furthermore, it is important to use the same buffer for filling the electrophoresis tank and gel preparation. Using two different batches or types of buffer may lead to variations and compromise the quality of the results.
Advantages of Agarose Gel Electrophoresis
There are various advantages to using agarose gel electrophoresis in molecular biology research. Some of the benefits of agarose gel electrophoresis include:
- Accuracy: Agarose gel electrophoresis is a dependable and precise method for sorting DNA fragments based on their size. It enables scientists to determine the presence or absence of specific DNA fragments and estimate their size range.
- Reliability: Agarose gel electrophoresis is a commonly used and well-established technique in molecular biology. It has been thoroughly tested and is regarded as a reliable approach for DNA analysis.
- Cost-Effectiveness: When compared to other gel matrices used in electrophoresis, such as polyacrylamide gels, agarose is a cost-effective material. Agarose gel electrophoresis is a low-cost method for analysing DNA since it is commonly available.
- Reproducibility: When standardised techniques and quality control measures are followed, agarose gel electrophoresis is a highly reproducible technology. This ensures that results are consistent and comparable between studies and laboratories.
- Simple and Quick Preparation: Agarose gels are relatively easy and quick to make. They simply require a single-component agarose and do not require polymerization catalysts. The gel is simple to pour and solidifies at room temperature without denatureing the DNA samples.
- Sample Recovery: After separation, agarose gel electrophoresis enables for the recovery of DNA samples. The desired DNA bands can be excised from the gel for further downstream applications such as DNA purification, cloning, or sequencing.
These benefits make agarose gel electrophoresis an appealing option for a variety of DNA analysis applications. Its ease of use, low cost, accuracy, and reproducibility have all contributed to its widespread use in molecular biology research.
Disadvantages/Limitations of Agarose Gel Electrophoresis
While agarose gel electrophoresis is a popular technique with many uses, it does have some drawbacks. The following are some of the limitations of agarose gel electrophoresis:
- Concerns about safety: Agarose gel electrophoresis uses electrical current, potentially hazardous chemicals (such as ethidium bromide), and heating procedures, all of which can be dangerous if not handled appropriately. Adequate safety procedures and measures should be followed to safeguard the safety of researchers.
- Inaccurate Size Determination: Agarose gel electrophoresis only provides a rough estimate of DNA fragment sizes. The gel’s resolution is restricted, making it difficult to properly measure the size of DNA pieces. Accurate size measurement necessitates the use of additional procedures, such as DNA sequencing or other specialised techniques.
- Time-consuming: The agarose gel electrophoresis procedure can be time-consuming. Gel preparation, sample loading, electrophoresis, staining, and visualisation are all steps in the process. The entire process can take several hours or even a day to complete, depending on the experimental setup and the size of the DNA fragments being analysed.
- Manpower Requirement: Agarose gel electrophoresis frequently necessitates numerous researchers handling different procedures at the same time. With greater manpower, gel preparation, sample loading, and result interpretation can be more efficient. This might be a problem in laboratories with limited resources or employees.
- Limited Data: Agarose gel electrophoresis reveals the existence, size, and relative abundance of DNA fragments. It does not, however, offer sequence information. Additional techniques, such as DNA sequencing, are necessary for extensive sequence analysis.
When considering agarose gel electrophoresis as a technique for DNA analysis, it is critical to be aware of these limitations. If these constraints offer substantial hurdles to their research objectives, researchers should review their individual experimental requirements and consider alternative methodologies.
Applications of Agarose Gel Electrophoresis
As a flexible technique for separating nucleic acid fragments, agarose gel electrophoresis has several applications in molecular biology and related sciences. Among the most important applications are:
- DNA Fragment Analysis: Agarose gel electrophoresis is a popular method for determining the size and number of DNA fragments. It is used to evaluate the effectiveness of DNA extraction, PCR amplification, and DNA modification investigations.
- RNA Analysis: RNA samples, including mRNA and tiny RNA molecules, are studied using agarose gel electrophoresis. It aids in the assessment of RNA integrity, the analysis of gene expression patterns, and the investigation of RNA processing events.
- Purification of DNA Fragments: Agarose gel electrophoresis is used to separate specific DNA fragments from complicated mixtures. Following separation, desirable DNA bands can be cut from the gel and purified for further use.
- Genetic Engineering: In genetic engineering experiments, agarose gel electrophoresis is required. It allows for the confirmation of successful DNA cloning, the detection of specific DNA fragments, and the evaluation of plasmid quality.
- DNA Fingerprinting: DNA fingerprinting techniques such as restriction fragment length polymorphism (RFLP) analysis and short tandem repeat (STR) analysis rely heavily on agarose gel electrophoresis. It aids in the separation of DNA fragments of varying lengths, enabling for the comparison and identification of distinct genetic profiles.
- Genotyping: Genotyping investigations use agarose gel electrophoresis to identify genetic variants and mutations. It allows for the examination of DNA markers such as single nucleotide polymorphisms (SNPs) or microsatellites for population genetics, disease association research, and forensic investigations.
- DNA Sequencing: In the early stages of DNA sequencing operations, agarose gel electrophoresis is used. It aids in the verification of the presence of PCR products and the quality of DNA libraries prior to the sequencing stage.
- Analysis of PCR results: Agarose gel electrophoresis is used to analyse PCR results and confirm successful amplification. It enables visualisation and verification of expected DNA fragment sizes.
- Teaching and Education: Agarose gel electrophoresis is commonly used in educational settings to expose students to molecular biology principles. It allows students to have hands-on experience with DNA separation and analysis.
- DNA Library Construction: Agarose gel electrophoresis is employed to verify the quality and quantity of DNA fragments in constructing genomic or cDNA libraries.
- Southern Blotting: Agarose gel electrophoresis is an integral part of Southern blotting, a technique used to detect specific DNA sequences. After electrophoresis, the DNA fragments are transferred to a membrane and hybridized with labeled DNA probes.
- Northern Blotting: Similar to Southern blotting, Northern blotting involves the separation of RNA fragments on an agarose gel, followed by their transfer to a membrane for detection of specific RNA molecules.
- Western Blotting: Although primarily used for protein analysis, Western blotting may also involve agarose gel electrophoresis for the separation of protein samples based on size before transfer onto a membrane.
- Protein Analysis: Agarose gel electrophoresis can be used for the separation and analysis of proteins, particularly for native gel electrophoresis, where proteins are separated based on their size, shape, and charge under native conditions.
- Isolation of DNA Fragments for Cloning: Agarose gel electrophoresis is employed to isolate specific DNA fragments of interest from the gel for subsequent cloning and other molecular biology techniques.
- Confirmation of Gene Knockout or Transgene Insertion: Agarose gel electrophoresis can be utilized to confirm the successful knockout of a gene or the insertion of a transgene in genetic engineering experiments.
- Quality Control in DNA/RNA Synthesis: Agarose gel electrophoresis is used for quality control checks during DNA and RNA synthesis, ensuring the correct length and integrity of the synthesized molecules.
- Microbial Identification: Agarose gel electrophoresis, along with techniques like PCR, is used for the identification and differentiation of microbial species based on their DNA profiles.
- Environmental DNA Analysis: Agarose gel electrophoresis can be employed to analyze environmental DNA (eDNA) samples to identify species present in a particular environment based on their DNA fragments.
Optimization of Agarose Gel Electrophoresis
To optimize agarose gel electrophoresis, you can consider the following recommendations:
- Buffer Selection: It is preferable to use TAE (Tris-Acetate-EDTA) buffer whenever possible, as the Borate in TBE (Tris-Borate-EDTA) buffer can sometimes create problems. TAE buffer provides good resolution and efficient separation of DNA fragments.
- Optimal Current Input: Follow the protocol or manufacturer’s recommendations for running the gel at the appropriate current input. Running the gel at faster or slower speeds than recommended can result in DNA smearing or diffusion within the gel, respectively.
- Gel Concentration: Use a 0.8% agarose gel for genomic DNA samples and a 1.8% agarose gel for PCR fragments. Adjusting the gel concentration can optimize the separation of DNA fragments of different sizes.
- Avoid Buffer Reuse: It is essential not to reuse the electrophoresis buffer as it may contain contaminants or altered pH that can affect the reproducibility and accuracy of results. Prepare fresh buffer for each gel run.
- Gel Disposal: After use, it is important to discard the gel and not reuse it. Reusing a gel can introduce contaminants and affect the resolution and integrity of DNA bands.
- Voltage Optimization: Running DNA at higher voltages can generate excessive heat, leading to gel melting and degradation of DNA samples. Follow the recommended voltage settings to ensure proper separation without compromising DNA integrity.
- Check for Bubbles: Before pouring the gel, carefully inspect it for the presence of bubbles. If bubbles are observed, it is advisable not to use that gel, as they can interfere with the migration of DNA fragments.
- Intact Wells: Ensure that the wells of the gel are intact and not broken before pouring the sample. Broken wells can lead to sample leakage and compromised results.
- Agarose Powder Selection: The personal preference mentioned for Seakem agarose powder is subjective, but cost-effective options that yield reliable results can be explored. Choose an agarose powder that suits your budget and provides the desired resolution and quality of DNA separation.
By following these optimization tips, you can enhance the quality and reliability of your agarose gel electrophoresis results. It’s important to note that these recommendations may need to be adjusted based on the specific requirements of your experiment and the recommendations provided by the manufacturer of the gel and related materials.
Precautions
Certain precautions must be taken when performing agarose gel electrophoresis to ensure safety and accurate findings. Consider the following crucial precautions:
- Personal Protective Equipment: When working with agarose powder, agarose gels, and other chemicals in the procedure, always use gloves, face masks, and goggles. This helps to safeguard against potential risks.
- Heat Protection: It is critical to wear oven gloves (heat resistant) during the boiling step when making the agarose gel. Plastic gloves should not be used for this stage because they can melt and cause burns.
- Handling Ethidium Bromide (EtBr): EtBr is often used to stain DNA in agarose gels. It is a dangerous chemical that is both carcinogenic and mutagenic. When handling EtBr, take the essential precautions, such as working in a well-ventilated location, utilising a fume hood if one is available, and wearing proper protection gear.
- Proper Disposal: Follow the applicable safety recommendations and local requirements for disposing of agarose gels and any hazardous materials, such as EtBr-contaminated solutions. Follow the recommended hazardous waste disposal procedures.
Concerning the topic of developing an electrophoresis device in your laboratory, it is not recommended unless you have the appropriate experience and resources. To ensure accurate and safe separation of DNA fragments, commercially available electrophoresis equipment is developed with safety measures and precise specifications. To provide reliable results and minimise potential dangers connected with handmade or improvised equipment, it is recommended to utilise commercially designed electrophoresis instruments.
When working with potentially dangerous materials and equipment, laboratory safety is critical. To keep yourself and others safe in the lab, always follow proper safety measures and rules.
FAQ
Q. In gel electrophoresis what is the agarose used for? / why is agarose used in gel electrophoresis?
Agarose gels are typically used to visualise fragments of DNA. The concentration of agarose used to make the gel depends on the size of the DNA fragments you are working with. The higher the agarose concentration, the denser the matrix and vice versa.
Agar has a lot of sulphate groups (sulfur surrounded by oxygens). These are also negatively-charged, so they can interfere with how the DNA moves through the gel. So it would make a bad matrix for electrophoresis. BUT agarose is neutral, making a good matrix for electrophoresis.
Q. How are dna fragments separated on an agarose gel during electrophoresis?
To separate DNA using agarose gel electrophoresis, the DNA is loaded into pre-cast wells in the gel and a current applied. The phosphate backbone of the DNA (and RNA) molecule is negatively charged, therefore when placed in an electric field, DNA fragments will migrate to the positively charged anode.
In gel electrophoresis what is the agarose used for?
In gel electrophoresis, agarose is used to create a solid gel matrix that serves as the medium for separating biomolecules, such as DNA or proteins, based on their size.
Agarose is a polysaccharide derived from seaweed and is commonly used in molecular biology and biochemistry research. When agarose powder is mixed with a buffer solution, typically Tris-acetate-EDTA (TAE) or Tris-borate-EDTA (TBE), and heated, it dissolves and forms a homogeneous gel mixture. As the mixture cools down, the agarose molecules reassociate and form a gel network.
The gel matrix created by agarose has a porous structure with small tunnels or pores through which biomolecules can migrate. The size of the pores in the gel is determined by the concentration of agarose used. Higher agarose concentrations result in smaller pores, while lower concentrations produce larger pores.
During gel electrophoresis, the agarose gel is placed in an electrophoresis chamber and submerged in a buffer solution that conducts electricity. Electrodes are connected to the ends of the chamber to create an electric field. When an electric current is applied, charged biomolecules, such as negatively charged DNA or proteins, migrate through the gel matrix.
The agarose gel acts as a molecular sieve, where smaller biomolecules can move more easily through the gel matrix, while larger biomolecules encounter more resistance and migrate more slowly. As a result, the biomolecules separate into distinct bands within the gel according to their sizes. These bands can be visualized by staining the gel with DNA-specific dyes or protein stains.
In summary, agarose is used in gel electrophoresis to create a solid gel matrix with pores that facilitate the separation of biomolecules based on size during the application of an electric field.
How does agarose gel electrophoresis work?
Agarose gel electrophoresis is a widely used technique in molecular biology and biochemistry to separate and analyze DNA or RNA fragments based on their size. The process involves the movement of charged nucleic acid molecules through a gel matrix in response to an electric field.
Here’s a step-by-step explanation of how agarose gel electrophoresis works:
Gel Preparation: Agarose, a polysaccharide derived from seaweed, is mixed with a buffer solution and heated to form a gel. The concentration of agarose can be adjusted to create gels with different pore sizes, which will affect the separation of nucleic acid fragments. The gel is typically cast in a rectangular mold with wells at one end for sample loading.
Sample Preparation: The DNA or RNA samples to be analyzed are mixed with a loading buffer that contains a tracking dye, such as bromophenol blue or xylene cyanol, which helps visualize the movement of the samples during electrophoresis. The samples are denatured by heating, which breaks the double-stranded DNA into single strands.
Loading Samples: The denatured DNA or RNA samples are carefully pipetted into the wells of the gel. It’s important to handle the gel and samples gently to avoid damaging the gel matrix and causing distorted migration patterns.
Electrophoresis: The gel is submerged in a buffer-filled electrophoresis chamber, and electrodes are placed at each end of the chamber. The DNA or RNA molecules in the samples carry a net negative charge due to the phosphate backbone, so they migrate towards the positive electrode (anode) when an electric current is applied.
Separation: As the electric current passes through the gel, the DNA or RNA molecules begin to move through the pores of the gel matrix. Smaller fragments move more quickly and travel farther, while larger fragments experience more resistance and move more slowly. This separation is based on the size-to-charge ratio of the molecules.
Visualization: After a specific amount of time, the electrophoresis is stopped, and the gel is removed from the chamber. The separated DNA or RNA fragments are not immediately visible, so a staining method is used to visualize them. Ethidium bromide or other DNA-specific dyes are commonly used for staining. The dye intercalates between the DNA bases and fluoresces when exposed to ultraviolet light.
Analysis: The stained gel is examined under UV light, and the separated DNA or RNA fragments appear as distinct bands of varying intensities. The distance traveled by each fragment can be compared to known size markers (DNA ladder) loaded in separate wells to estimate the size of the unknown fragments.
By analyzing the resulting gel, researchers can determine the size distribution of nucleic acid fragments in a sample, identify specific fragments of interest, or confirm the success of DNA amplification (e.g., PCR).
I like the interpretation