What is Semiconductor Sequencing?
Semiconductor sequencing is the integration of microchip technology with DNA sequencing where electrical signals replace the detection of light. This sequencing method is unique because it uses complementary metal-oxide semiconductor chips associated with sequencing by synthesis chemistry; so, in this respect, it opens up opportunities to make DNA analysis quicker and less expensive than its classical counterparts.
DNA sequencing used to depend a lot on the chain-termination method. Many new sequencing methods have concentrated on detecting light signals. However, semiconductor sequencing works differently. It finds electrical changes that happen when protons are released during DNA building—removing the need for complicated optical systems.
This method was first sold to the public in 2010 through Ion Torrent sequencing, which was started by Jonathan Rothberg. His research had already helped create earlier sequencing tools, like the Roche 454 system. Later, Ion Torrent Systems became a part of Thermo Fisher Scientific.
Why is it different? Traditional sequencing approaches typically require a lot of equipment to detect light. Semiconductor sequencing has made this much easier. Electrical detection replaces large, expensive optical setups, making the process easier to use.
This sequencing technology has not only made the process faster but also opened new avenues in DNA research. Combining the latest microchip technology with biochemical processes has changed how genetic data are collected.
What is the working Principle of Semiconductor Sequencing?
It operates on the principle of sequencing by synthesis (SBS) and uses ion-sensitive field-effect transistors (ISFETs). The sensors are made on CMOS chips, and they detect the electrical changes during nucleotide incorporation.
It essentially captures the hydrogen ions (H⁺) released during DNA polymerisation. The process is both simple and efficient. Each sequencing chip has a dense array of wells and pH sensors, which allows for parallel processing of DNA fragments.
First, DNA pieces are prepared. The fragments with adapters are copied using emulsion PCR (emPCR). In this step, the fragments are surrounded by beads and PCR materials in an oil mix. After copying, the beads with DNA are taken out and put into the chip wells.
Sequencing starts as nucleotides are added one after the other. In case a nucleotide matches the DNA template, it gets added to the strand. A hydrogen ion is liberated. These ions shift the pH in the well and cause a shift in electric potential.
The ISFET sensors below the wells sense these movements. Information created from such electrical changes is then processed to determine which nucleotide was added. There is no requirement for optical systems, thus reducing the complexity and cost of the process.
Combining biochemical reactions with microchip technology, this is a revolutionary advancement in sequencing techniques.
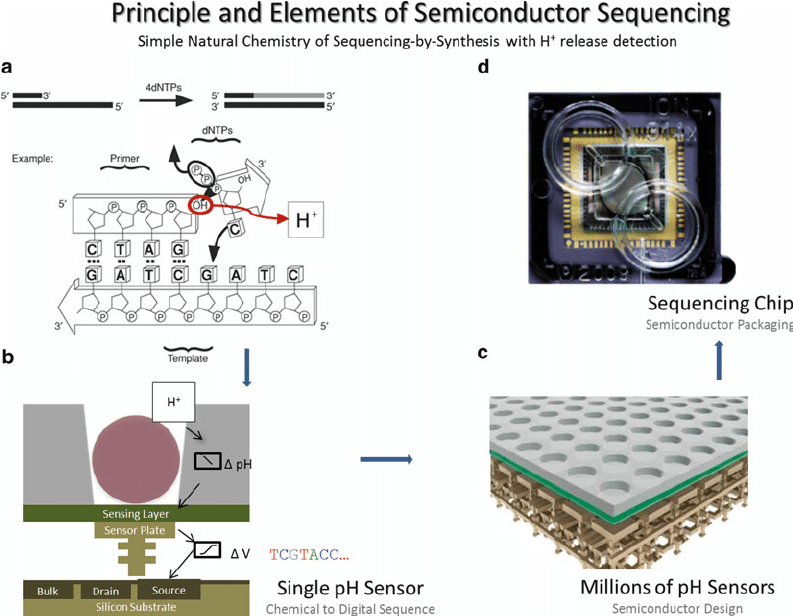
What sensors and technology is used in Semiconductor Sequencing?
- There is intense reliance by semiconductor sequencing on two major technologies, namely CMOS chips and ISFET sensors. Along with these, many years of study into semiconductor devices and methods of DNA sequencing come into play to facilitate this sequencing process.
- These are extremely important chips as far as the system is concerned, since CMOS chips, manufactured first during the 1970s, have become necessary for most present electronic devices that range from cell phones to digital cameras. Semiconductor sequencing allows a million sequencing reactions on one chip alone, thanks to the use of CMOS technology. It does so much more rapidly and cost-effectively when optical detection is eschewed for this method.
- The ISFET sensor is another significant sensor. This is an electrochemical sensor with a very high sensitivity in detecting even the smallest changes in ion concentration. Such sensitivity is required to detect the release of hydrogen ions during DNA polymerisation.
- ISFETs can easily work with CMOS chips, allowing many sensors to be placed on one chip. This compatibility is a big benefit because it enables fast sequencing without needing large optical systems.
- The invention of CMOS-based sensors and ISFET technology has improved DNA sequencing. The sensors are small, inexpensive, and can be easily integrated with other technologies to bring about great improvements in sequencing. This has made DNA sequencing easier and less expensive.
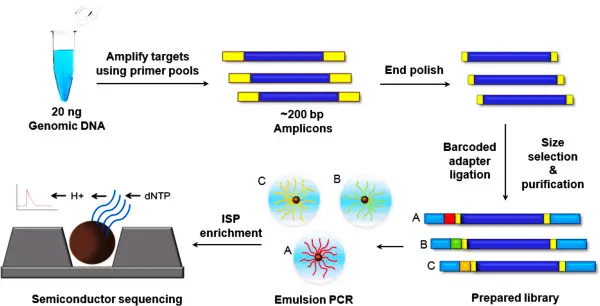
What is the process involved in semiconductor sequencing and how does it work?
Sequencing semiconductors is a very fast and complicated process. It has several steps that build on top of each other and are all necessary to get true genetic data. Not only does the process use basic DNA sequencing techniques, it also adds some new features that make it different from other ways.
- The first step is to get the library ready. Genomic DNA is taken from the sample and broken up. After this, adapter sequences are added to both ends of the DNA that has been broken up. These adapters make sure that the DNA is ready for the decoding process.
- After the library is ready, the DNA pieces need to be made bigger. An droplet PCR is used to do this. Ion Sphere Particles (ISPs) are small beads that help carry the DNA. The DNA pieces stick to them. The ISPs are put in a mixture of water and oil, which makes tiny reactors. DNA can be copied into millions of copies in this controlled setting. Isolated, cleaned, and ready for the next step are only the beads whose DNA has been properly amplified.
- It is in the ordering step where the real magic happens. The ISPs that have been made are carefully put into the semiconductor sequencing chip, which has millions of microwells. One ISP is in each well. A polymerase enzyme adds nucleotides to the DNA template when sequencing starts. The pH in the well changes slightly because of this process, which produces hydrogen ions. These changes in pH are picked up by the ISFET sensor at the bottom of the microwell. The electrical output that is made is then handled and changed into voltage, which shows which nucleotide has been added.
- After the sequencing is done, the raw data goes through a very important part of data analysis. Nucleotide patterns are made from the signals that are made during sequencing. After that, these genes are put through a number of quality control checks and are matched up with reference genomes. This step makes sure the data is correct and ready to be looked at more closely. Bioinformatics tools that are very advanced help put together the DNA code, which can then be analyzed and understood to find biological meaning.
- Traditional methods and new ideas are combined in the process of semiconductor sequencing, which creates a quick and accurate way to decode genes. Because it works so well, this technology is not only important for study, but also for healthcare uses where speed and accuracy are very important. In spite of the fact that the field is always changing, the basic steps of semiconductor sequencing will probably be important for many years to come.
What key advantages or benefits does semiconductor sequencing offer compared to other sequencing methods?
- One important benefit semiconductor sequencing has for several uses is it is quite cost-effective. Utilizing CMOS technology helps save on the two main aspects in terms of producing and operating expenses. Considering most traditional sequencing approaches, costly optical systems often involve optical instrumentation and, in most cases, higher-priced light source equipment compared with the silicon-based pH sensors involved in semiconductor sequencing.
- This technology also gets rid of complicated systems of optics. Thus, the sequencing process is less cumbersome and more straightforward. Another advantage is that it uses native, unaltered nucleotides as opposed to chemically altered or fluorescently labeled bases, meaning that it makes the whole process simpler in general.
- Another advantage is the real-time ability to detect ions. Changes in pH when nucleotides are added to the DNA strand can be observed using semiconductor sequencing. This is very helpful for clinical tests and finding pathogens because fast results are very important in these cases.
- Semiconductor sequencers are fast, small, and easy to use. They are not like optical-based systems, which are usually big and hard to manage. Semiconductor sequencing technology gets rid of the need for heavy equipment. This makes the system easier to carry and good for labs with little space.
- Semiconductor sequencing is easier to use. The system requires less training than more complex platforms, making it accessible to more users. Easy use, along with the lower costs, makes it more attractive for regular sequencing tasks.
What are the main limitations or drawbacks that researchers face when using semiconductor sequencing technology?
- Semiconductor sequencing has pros and cons. Shorter read lengths than other sequencing techniques are a downside. Shorter reads can make complicated genome assembly harder, requiring more data processing.
- This sequencing process requires extensive data analysis tools and skills. Mistakes might arise when interpreting results without the right tools and understanding.
- Homopolymer areas are another issue. These stretches of successive identical bases are difficult for semiconductor sequencing to read. In genomes with repeated sequences, this might cause erroneous base calling.
- Environmental factors and buffer systems can affect pH detection accuracy. Even modest changes in these variables might produce inconsistent measurements and base calling mistakes.
- In repetitive and GC-rich areas, semiconductor sequencing may have a greater error rate despite its speed and simplicity. In precision-critical applications, this makes it less trustworthy.
In what fields and applications is semiconductor sequencing currently being used?
- Semiconductor sequencing has several uses. It is commonly used in clinical diagnostics to discover genetic changes connected to illnesses, including cancer. Detecting mutations helps personalize treatment options.
- This method helps cancer researchers study cancer-specific mutations and tumor genetics. This helps explain cancer progression.
- The technique is used to diagnose infectious disorders, especially pathogens. It can identify infection causes by sequencing microbial genomes, providing vital therapeutic information.
- Semiconductor sequencing is also popular in microbiome research. It enables researchers examine gut microbiomes and microbial populations in different situations. This is crucial to understanding bacteria in health and the environment.
- Semiconductor sequencing is crucial in agriculture. Plant and animal genome sequencing aids agricultural improvement and breeding. This boosts disease resistance and yield.
- For targeted DNA and RNA sequencing, semiconductor sequencing works. It helps discover genetic disease variants in exome sequencing.
- ChIP-Seq maps protein-DNA interactions using the technique. This illuminates epigenetic changes that alter gene expression and cell function.
- For single-cell transcriptomics, semiconductor sequencing is used to analyze gene expression. When studying ultra-small tissue samples, this helps researchers understand gene activity.
- Finally, microbial genome sequencing is efficient. Its fast processing speed and low cost make it suitable for large-scale microbial genome sequencing, including bacteria and viruses.
- Semiconductor sequencing is used for targeted and amplicon sequencing in research and diagnostics. Researchers can focus on genomic areas or DNA fragments using these methods to get precise data.
- https://en.wikipedia.org/wiki/Ion_semiconductor_sequencing
- https://www.thermofisher.com/us/en/home/life-science/sequencing/next-generation-sequencing/ion-torrent-next-generation-sequencing-technology.html
- https://academic.oup.com/bioinformatics/article/31/8/1199/212680
- https://www.semicontaiwan.org/en/node/10101
- https://microbenotes.com/semiconductor-sequencing/
- https://pmc.ncbi.nlm.nih.gov/articles/PMC4024476/
- https://www.illumina.com/science/technology/next-generation-sequencing/sequencing-technology/semiconductor-sequencing-cmos.html
- https://sapac.illumina.com/science/technology/next-generation-sequencing/sequencing-technology/semiconductor-sequencing-cmos.html
- https://www.nature.com/articles/nature10242