DNA replication is the process by which genetic material is copied before cell division happens, it’s considered essential for inheritance.
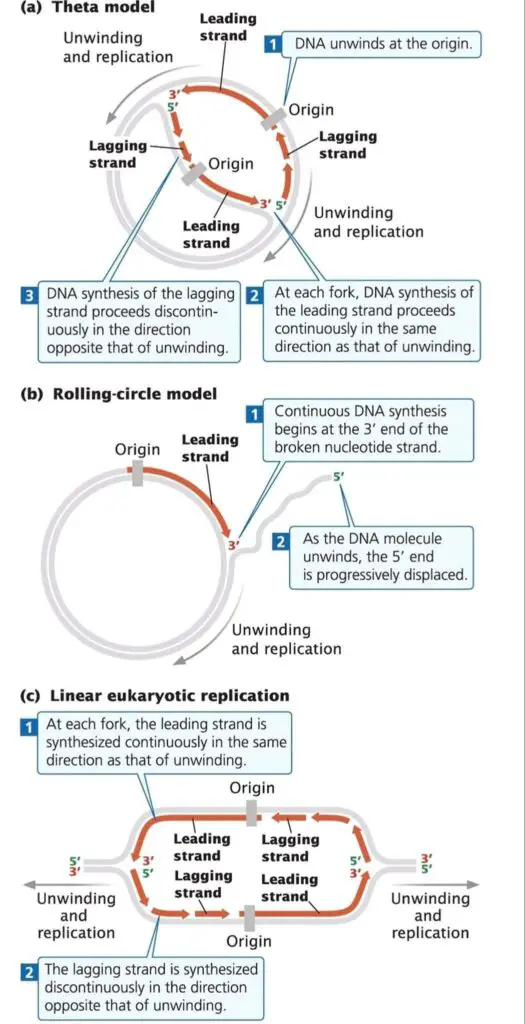
Among different models, the main ones observed are Theta model, Rolling Circle model, and Linear DNA replication.
Each model are used by different organisms or DNA types (like plasmids, viruses, or eukaryotic chromosomes).
- Theta model of replication was mainly seen in E. coli and other circular DNA molecules.
- It’s called “Theta” because during replication, the circular DNA takes a shape similar to greek letter θ (theta).
- In this model, replication begins at a specific site called origin of replication (oriC), where strands get separated.
- As replication forks move in both directions, the structure appears as a double-stranded loop— kind of like a bubble forming in the circle.
- DNA polymerase enzyme is used for adding nucleotides to new strands, and both strands acts as templates at the same time.
- The process may be bidirectional or sometimes unidirectional, depending on the organism and replication origin properties.
- After replication completes, two identical circular DNA molecules are produced / separated.
- Walking down the replication fork, the new DNA strands were elongated while parental strands unwind.
- Rolling Circle model is found mainly in bacteriophages (like ΦX174, λ phage), plasmids, and some viruses.
- In this type, one of the DNA strands is cut at a specific point by an endonuclease enzyme.
- The 3’-OH end that’s produced works as a primer for DNA polymerase to start synthesis.
- The old strand (template) is displaced as replication continues around the circle again and again.
- This displaced single-strand DNA (ssDNA) can later serve as a template for making its complementary strand.
- The process seems kind of continuous / rolling, so that’s why it’s named Rolling Circle.
- Often, long concatemeric DNA (having several genome copies linked in series) are formed before being cut into individual units.
- It has been noticed that replication through this model is faster than the theta-type.
- Sometimes multiple rounds of replication happen simultaneously on the same molecule, which make it very efficient for producing copies quickly.
- The enzymes involved are DNA polymerase, ligase, and sometimes helicase-like proteins that unwind the template.
- Linear DNA replication occurs in eukaryotes and in some viruses that have linear DNA instead of circular.
- In these cases, replication begins at multiple origins along the DNA molecule, not just one origin like in circular DNA.
- From each origin, replication forks move outward bidirectionally forming many replication bubbles at once.
- The lagging strand synthesis needs short RNA primers that are later removed and replaced by DNA, it’s a semi-discontinuous process.
- Because linear DNA has ends, special structures called telomeres and enzyme telomerase are required to prevent loss of DNA ends during replication.
- Without telomerase activity, the chromosome ends would get shortened each cell division – leading to aging or genome instability in cells.
- Each replication unit is completed independently, and later all are joined forming the complete chromosomal DNA.
- Such as, in human cells nearly 30,000–50,000 replication origins may be active during S-phase.
- The coordination of initiation and termination are regulated by cell cycle proteins and checkpoints.
The three models, though different in structure and mechanism, shares the same fundamental idea – that DNA must be copied accurately before cell division.
In prokaryotes, theta replication is more common, while eukaryotes relies on linear model; viruses/plasmids often use rolling circle.
Replication models are selected by organisms depending on the DNA topology, enzymatic system, and replication purpose.
Sometimes overlapping features occur, for instance, some plasmids may shift from theta to rolling circle under certain conditions.
Overall, the diversity in replication strategies shows how evolution have modified the DNA duplication process for various biological needs.
And that’s basically how these different DNA replication models—Theta / Rolling Circle / Linear—are overviewed in molecular biology.
1. Rolling circle replication
Rolling circle replication is a type of DNA replication seen mostly in small circular DNA molecules, like plasmids and certain viruses (such as ΦX174 or λ phage).
It’s considered a fast and efficient process, by which many copies of DNA can be produced from a single circular molecule.
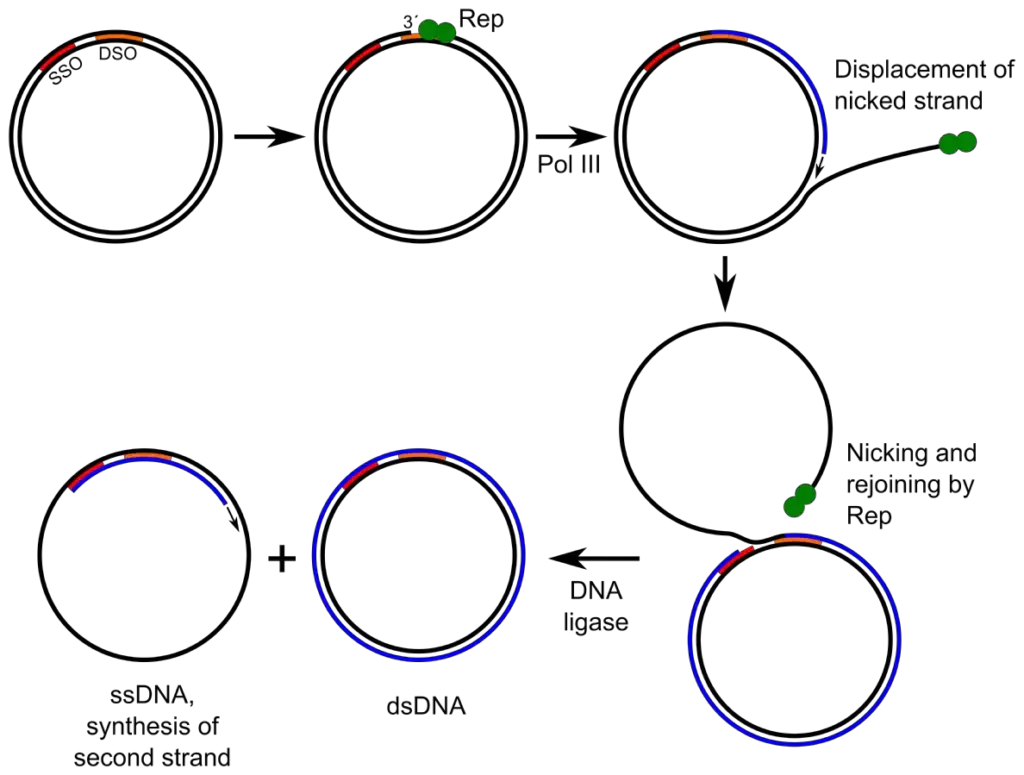
In this method, one strand of the circular DNA is cut by an enzyme called endonuclease, at a specific point known as the origin of replication.
After being nicked, the free 3’-OH end acts as a starting site for DNA polymerase to begin synthesis of a new strand.
The intact strand serves as a template, while the old (displaced) strand is pushed out as the new one is made.
As replication continues, the template keeps rotating / rolling—hence the term “rolling circle.”
The displaced single-strand DNA (ssDNA) may later be converted to double-stranded form by synthesis of a complementary strand.
The new strand elongates continuously in 5’→3’ direction, while the old one gets displaced progressively.
Usually, long linear DNA molecules containing multiple genome copies (called concatemers) are formed during this mechanism.
Such concatemers are later cleaved and circularized to form individual DNA units, mostly by ligase enzyme activity.
In plasmids, the process is used for rapid DNA transfer / amplification during conjugation or replication inside bacterial cells.
The initiation step is dependent on specific rep proteins, which recognizes and nicks the double-stranded DNA at ori (origin site).
Sometimes multiple replication events happen on same molecule simultaneously—it makes the process quite productive.
The replication fork moves around the circle like a wheel turning on its axis, producing a tail of single-stranded DNA behind.
After completion, the parental strand remains circular, and the newly synthesized strand is released or converted into double-stranded DNA.
Walking down the molecular events, the replication looks continuous but it actually involves discontinuous synthesis for complementary strand formation.
The enzymes mainly involved are DNA polymerase, DNA ligase, and endonuclease (or nickase).
Rolling circle replication is not found in all organisms; it’s typical of certain phages, plasmids, and sometimes mitochondrial DNA of yeasts.
Such as, in Staphylococcus aureus plasmid pT181, this mechanism was well documented.
The model provides a clear example of how a circular DNA can be multiplied without a need of complete unwinding of both strands.
Errors in nicking or ligation may result in incomplete replication, sometimes leading to strand breaks or concatemer instability.
The advantage of this model lies in its simplicity and the ability to rapidly produce many DNA copies using minimal enzymatic machinery.
For viruses, it’s beneficial since they can replicate their genomes quickly after infection inside host cells.
The whole mechanism happens inside cytoplasm or nucleus, depending on the type of organism and DNA molecule.
And that’s basically how Rolling Circle replication operates—like a wheel that keeps turning, copying the DNA again and again until it stops.
Steps of Rolling circle replication
- Rolling circle replication is initiated at a specific origin (ori), and the event is recognized by rep proteins, which bind and prepare the site.
- The double-stranded circle is then nicked by an endonuclease (nickase), producing a free 3’-OH and a 5’-phosphate, and the nicking is performed at a defined position.
- From the 3’-OH end, DNA polymerase is engaged and synthesis is begun, while the 5’ end is displaced progressively.
- The intact strand is used as template, and complementary bases are added to the growing 3’ end, in a continuous 5’→3’ extension.
- As synthesis proceeds the circular template is caused to rotate / roll, which results in the displaced strand forming a single-stranded tail (ssDNA).
- The displaced ssDNA remains attached transiently to the circle, or it may be released entirely, depending on the system.
- Short RNA primers are laid down on the displaced ssDNA to initiate synthesis of its complementary strand, by a primase or priming protein.
- Complementary strand synthesis is then carried out, and Okazaki-like fragments are sometimes made on the complementary strand (lagging synthesis).
- The newly synthesized complementary sequences are joined by DNA ligase, producing double-stranded product (dsDNA) from the displaced strand.
- Long linear concatemers (several genome copies linked end-to-end) are often produced when continuous rolling continues, and these are seen as replication products.
- Concatemers are cleaved at specific sites, and individual genome units are circularized; ligation is performed to seal nicks.
- The original parental circle is usually left intact, it can be reused as a template for further rounds, and sometimes multiple initiations occur on same circle.
- Rep proteins (or Rep) are often required for both nicking and for termination/ligation, and they are essential in plasmid systems.
- In some bacteriophages, packaging proteins recognize concatemer junctions and cut during packaging, so the genomes are inserted into capsids directly.
- The enzymes that are mainly required are DNA polymerase, ligase, endonuclease (nickase), and in many systems helicase-like proteins help unwinding.
- Strand displacement is continuous for the leading new strand, yet discontinuous events are involved for complementary strand—this mixed mode is characteristic.
- Walking through the reaction, errors at nicking or ligation are sometimes produced, leading to incomplete molecules or breaks, which may be repaired or degraded.
- Such as, in Staphylococcus aureus plasmid pT181, RepC is used to nick and initiate rolling, which was documented experimentally.
- The speed of rolling circle replication is often higher than theta-type, so rapid genome amplification is achieved, which benefits phage replication and plasmid spread.
- Multiple tails (ssDNA) may be synthesized from a single circle simultaneously, when multiple initiations are allowed, making it highly productive.
- The displaced ssDNA is converted to dsDNA after priming and synthesis, or it is packaged as ssDNA in some viruses (for example, ssDNA phages).
- The whole process is dependent on precise cleavage and rejoining; failure to ligate correctly will leave nicks, and the DNA integrity are compromised.
- In bacteria, the replicated plasmid can be transferred by conjugation after replication, and plasmid copy number is increased by such rolling events.
- Fragmented notes: Nick, synthesize, displace, prime, ligate.
- The mechanism is versatile, yet simple, and it is favored when quick, multiple-copy production is required; it’s used by plasmids, some phages, and even mtDNA (in some yeasts).
- Errors and variations are tolerated to an extent, they are corrected by repair pathways or cause instability if persistent.
- That’s the stepwise outline of circular (rolling) replication — continuous leading synthesis, displaced tail formation, primer-dependent lagging conversion, concatemer formation and cleavage into circles.
Advantages of Rolling circle replication
- High replication speed is often enabled by rolling circle replication (RCR), because a single nick at the origin is made and continuous DNA elongation is permitted, producing long concatemer(s) rapidly, which is why rapid amplification is favored.
- Resource efficiency is observed, as only one primer/nick is required and less initiation machinery is demanded, so host factors are spared and replication is economical for small plasmids / phage genomes.
- Rapid production of copies is achieved, long ssDNA (single-stranded DNA) molecules are generated and later processed into unit-length genomes, facilitating packaging.
- Simplicity of mechanism is favored; strand-displacement synthesis is facilitated, replication forks are avoided, fewer complex replisome components are required and so the process is less topologically constrained.
- Adaptability is shown, the mechanism is exploited in bacteria, plasmids and some viruses (eg Escherichia coli plasmids, certain Geminiviridae), so versatility is high and mobile elements exploit it.
- Horizontal transfer is made easier, mobile genetic elements are amplified quickly, and thus propagation of plasmids/ phage is enabled, it often hijacks host enzymes (such as Rep protein/Rep).
Limitations of Rolling circle replication
- Error-correction is limited during extended single-stranded synthesis, mismatches are more likely to persist, hence mutation rate can be elevated and genetic variability is increased.
- Secondary structures (hairpins, stems) in long ssDNA are frequently formed, causing polymerase stalling or template-switching, interruption of synthesis and thus fidelity and continuity may be compromised.
- Concatenated products must be processed (nicking/ cleavage / ligation) to yield unit-length genomes, and if processing enzymes (endonucleases/ ligases) are absent or inefficient, genome maturation is impaired.
- Size-constraints are imposed; very large genomes are not efficiently handled by RCR so it is mostly found in small replicons, plasmids and certain viruses rather than chromosomes — it’s a limit.
- Reliance on specific initiator proteins (eg Rep ) is required, when these proteins are mutated or inhibited, replication fails or is drastically reduced, host factors aside.
- Recombination between concatemers can lead to genomic rearrangements, deletions or insertions, instability may arise, and plasmid burden is sometimes increased on hosts; they sometimes replicate uncontrollably (copy-number variation) causing metabolic cost.
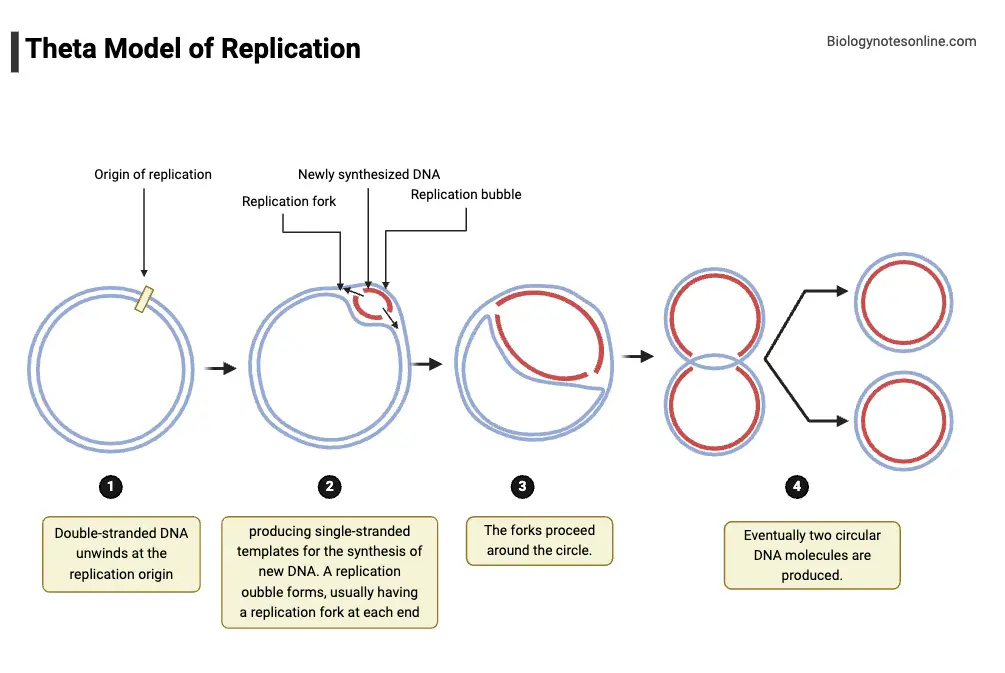
2. Theta Model of Replication
Theta model of DNA replication is described as a mechanism used by circular genomes, and it is commonly seen in E. coli and many plasmids.
The name “Theta” was given because the replication intermediate is visualized like the greek letter θ, when autoradiographs or electron micrographs are made.
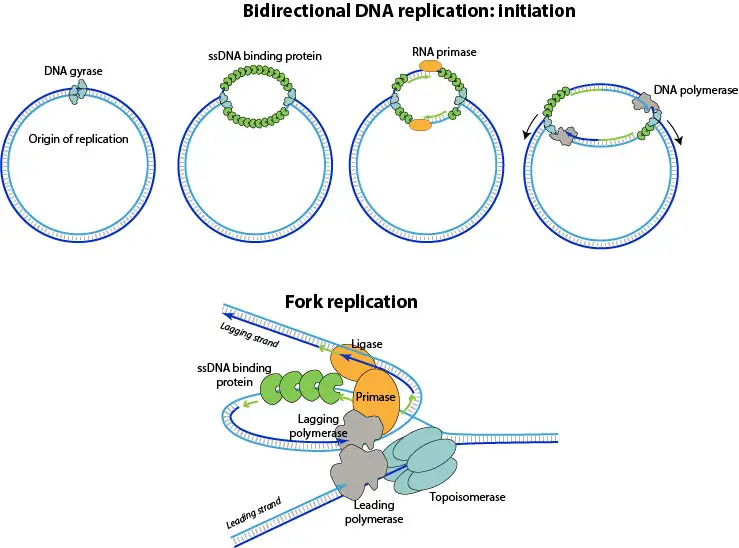
Replication is usually initiated at a single, defined origin (ori or oriC), where initiator proteins are bound and local unwinding is caused.
At the origin, the duplex DNA is unwound and a small replication bubble is formed, the bubble is stabilized by SSB proteins.
DnaA (in bacteria) is often the protein that is bound to oriC and it is used to open the duplex, and helicase is then loaded.
Helicase loading was followed by unwinding of DNA, and supercoils are relieved by DNA gyrase or topoisomerase II like enzymes.
Two replication forks are formed at the edges of the bubble, and they are moved (progressed) around the circle in opposite directions in many cases.
When forks move in both directions the process is called bidirectional replication, which speeds up copy-making for large circles.
On the leading strand synthesis is continuous and polymerase is attached, while on the lagging strand short Okazaki fragments are produced and later joined by DNA ligase.
RNA primers are laid down by primase, they are removed and replaced by DNA (DNA pol I), and nicks are sealed by ligase; this is the classic flow.
The whole reaction is carried out mostly in 5’→3’ direction, and proofreading functions are provided by polymerase exonuclease activities.
Replication forks may stall sometimes, and repair or restart pathways are engaged, causing delays or fork reversal, they are often problematic.
Termination is achieved when the two forks meet at terminus region (ter), where termination proteins (Tus in E. coli) may bind to block fork passage.
After termination the daughter circles are often interlinked (catenated), and decatenation is performed by topoisomerase IV or type II topoisomerases.
Each daughter circle is composed of one parental and one newly synthesized strand, showing the semi-conservative nature of copying.
Initiation is tightly regulated, and timing is controlled by DnaA levels and ATP/ADP status, which ensures replication starts only when allowed.
The efficiency and accuracy are high, but errors can be introduced, and mismatch repair pathways are used post-replicatively to correct them.
In some plasmids, theta replication was observed to switch modes or to be unidirectional depending on origin architecture, which is noted experimentally.
Walking along the process, the bubble was seen to enlarge as synthesis continues, and the θ-shape becomes obvious, a clear visual marker.
The process is preferred for moderate-to-large circular chromosomes because it maintains stability and control, rather than making long concatemers.
Such as, in E. coli, DnaA binds to DnaA-boxes at oriC and the local region is destabilized for helicase entry, experimental details were mapped.
The model is largely conserved among many prokaryotes, though variations in proteins and regulatory steps are present, and some species use different factors.
Simple advantages are provided: complete replication with proofreading is allowed, and correct segregation is facilitated by decatenation prior to cell division.
Fragmented note: origin → unwind → bubble → forks → synthesize (leading, lagging) → terminate → decatenate → two circles.
Steps of Theta Model of Replication
- Theta model of DNA replication is described as a mechanism used by circular genomes, and it is commonly seen in E. coli and some plasmids.
- Initiation is started at a defined origin (ori or oriC), where specific initiator proteins are bound and origin recognition is performed.
- DnaA (when present) is bound to DnaA-boxes at oriC, and local duplex destabilization is caused by its ATP-dependent action, which opens a small region.
- Walking into the origin, the strands were exposed, a small open complex is formed (this is unstable if SSBs are absent).
- Then, helicase (DnaB) is loaded onto single strands with help of loader proteins, and unwinding of duplex DNA is carried out as replication forks are set up.
- Single-strand binding proteins (SSBs) are attached to separated strands so that reannealing is prevented, and strand stability is maintained.
- Ahead of the fork, supercoils are relieved by DNA gyrase / topoisomerase activity, which is needed, otherwise torsional strain would accumulate.
- Primase is recruited and short RNA primers are synthesized, these primers provide 3’-OH ends for polymerase to act upon.
- DNA polymerase III is then engaged and synthesis is begun on both templates, new strands are elongated in 5’→3’ direction with proofreading.
- On the leading template, synthesis is continuous and is performed by the polymerase holoenzyme while it tracks with helicase.
- On the lagging template, synthesis is discontinuous, Okazaki fragments are produced, and primers are removed later; this is semi-discontinuous overall.
- RNA primers are removed by DNA polymerase I and the gaps are filled with DNA, and DNA ligase seals the nicks to complete backbone continuity.
- Two replication forks are formed at the bubble edges, and they are usually moved bidirectionally, so the replication bubble enlarges as synthesis continues.
- When forks progress they are coordinated yet each fork can stall independently, which invokes restart/repair pathways, and this may delay completion.
- Termination is effected when forks meet at terminus (ter) region, where termination sites and proteins (e.g., Tus in some bacteria) are bound to arrest fork progression.
- After completion, daughter circles are produced but they are often interlinked (catenanes), and decatenation is required for segregation.
- Type II topoisomerases (topo IV, or gyrase-like enzymes) are used to separate catenated circles by nicking-passing-resealing mechanism.
- Each daughter circle is composed of one parental and one nascent strand, which shows the semi-conservative nature of replication.
- Initiation events are tightly regulated by initiator protein levels, ATP/ADP status and methylation state at ori, control is exerted to permit only one round per cycle.
- The rate of replication is high (in bacteria about 1000 nt/sec is observed under optimal growth) and complete genome duplication is achieved within a defined time window.
- Errors are proofread by polymerase exonuclease activities and mismatches repaired post-replicatively by MMR pathways, but mistakes sometimes persist.
- The process is preferred for moderate-to-large circular chromosomes because fidelity and regulation are favorable, though theta is slower than rolling circle for copy-number bursts.
- Such as, in E. coli, DnaA-mediated origin opening is used, and Tus-ter interactions are exploited for orderly termination in some strains.
- Occasionally, unidirectional replication is observed in certain plasmids, and in those cases only one fork is active, which changes dynamics.
- Fragmented summary: origin recognition → unwinding → helicase loading → primer synthesis → bidirectional fork progression → Okazaki joining → fork meeting → termination → decatenation.
- The forks are moved by coordinated enzymatic complexes, they are occasionally impeded, and restart factors are recruited when problems arise.
- The model is visualized as a θ-shaped intermediate under EM, and that visual cue is often used to identify theta-mode replication.
- Overall, theta replication is carried out precisely, but it is not flawless, and cellular systems are relied upon to correct or resolve the occasional failures.
3. Replication of Linear Double-Stranded DNA (Bidirectional Replication)
Replication of linear double-stranded DNA is typically described for eukaryotic chromosomes, and it is carried out by multiple origins along each chromosome.
Initiation events are triggered at many origins of replication (origins), which are licensed and then activated in S-phase by initiator proteins.
Origin recognition complexes (ORC) are bound at origins in many eukaryotes, and origin firing is regulated by cyclin-dependent kinases and DDK; timing is controlled.
At each origin a replication bubble is formed after local unwinding, and two replication forks are established that move in opposite directions (bidirectional).
Helicase (MCM complex in eukaryotes) is loaded and activated, and unwinding of parental duplex is performed while supercoils are relieved by topoisomerase.
Single strands are stabilized by single-strand binding proteins (RPA in eukaryotes) to prevent reannealing, and they are held accessible for polymerases.
Primase (part of Pol α-primase in eukaryotes) synthesizes short RNA primers which are extended by DNA polymerases, providing 3’-OH ends for synthesis.
DNA polymerase ε is usually used for leading strand synthesis and DNA polymerase δ for lagging strand in many systems, though some flexibility is observed.
Leading strand synthesis is synthesized continuously toward the advancing fork, and the nascent strand is elongated in 5’→3’ direction with proofreading.
Lagging strand synthesis is discontinuous, Okazaki fragments are produced, and these fragments are later processed by removal of RNA primers and gap filling.
RNA primers are removed by nucleases (RNase H and FEN1) and replaced by DNA (Pol δ), and nicks are sealed by DNA ligase I to produce continuous strand.
Fork progression is coordinated by replisome complexes, but forks can stall, and restart pathways (template switching, translesion synthesis) are engaged when obstacles occur.
Termination is not at a single site for linear chromosomes; instead forks converge and are resolved when adjacent replicons meet, and processing of termini is required.
In organisms with linear chromosomes special end-replication problems are faced, because RNA primers cannot be replaced at extreme ends, leading to progressive shortening.
Telomeres (repetitive sequences) and telomerase enzyme are used in many eukaryotes to maintain ends, telomerase being active in germline, stem cells and some somatic cells.
The process is strictly regulated so that each origin is fired once per cell cycle, and re-replication is prevented by licensing factors that are inactivated after S-phase starts.
Replication speed varies: in eukaryotes forks often move at ~20–100 nt/sec (varies by cell type and conditions), while density of origins may be ~1 origin per 50–300 kb.
Okazaki fragments lengths differ: Such as, 100–200 nt in higher eukaryotes, and longer in some lower eukaryotes; processing enzymes adapt to fragment size.
Proofreading exonuclease activities of polymerases are relied upon to reduce errors, and mismatch repair (MMR) acts post-replicatively to correct remaining mismatches.
After replication, daughter chromatids are intertwined sometimes (catenanes) and are decatenated by type II topoisomerases prior to mitosis, allowing segregation.
The replicated chromatin is reassembled, nucleosomes are deposited (CAF-1, other chaperones) and epigenetic marks are re-established, which is crucial for function.
Coordination with cell-cycle checkpoints is enforced, and DNA damage responses may pause replication or activate repair, they ensure genome integrity.
In some viruses with linear dsDNA, bidirectional origins are used too, though host/viral proteins may be different, and packaging follows replication.
Walking through the overview: origins licensed → origin firing → bubble formation → bidirectional fork progression → leading/lagging synthesis → fragment processing → fork convergence → termination → decatenation → chromatin reassembly.
Fragment: origin → unwind → synthesize (leading, lagging) → process Okazaki → finish, simple.
The system is complex yet modular, it is regulated, adaptable, and mistakes are minimized but not eliminated, repair pathways are relied upon.
Steps of Bidirectional Replication
- Replication of linear double-stranded DNA is initiated at many origins which are licensed and marked, and ORC (Origin Recognition Complex) is usually bound at these sites.
- The origin licensing step is completed by loader proteins and factors, and the licensed origins are kept inactive until S-phase is entered.
- At S-phase start, origins are fired and origin activation is triggered by kinases (CDKs, DDK) so that helicase loading is converted into active unwinding.
- MCM helicase complex is loaded as an inactive double hexamer, and it is activated (converted) into two single hexamers that unwind DNA at the origin.
- Local unwinding is performed and a small replication bubble is formed, while single strands are coated by RPA (replication protein A) to prevent re-annealing.
- Primase (Pol α-primase) is recruited and short RNA primers are synthesized, these primers provide 3’-OH ends for extension.
- DNA polymerase ε is generally used for leading strand synthesis and DNA polymerase δ for lagging strand synthesis, though flexibility is observed.
- Leading strand synthesis is carried out continuously toward the fork, and high-fidelity extension is provided with proofreading exonuclease activity.
- Lagging strand is synthesized discontinuously as Okazaki fragments, and primer placement / fragment length are controlled by priming frequency.
- Okazaki fragments (100–200 nt in higher eukaryotes) are processed by nucleases (RNase H, FEN1) and gaps are filled by Pol δ, and nicks are sealed by DNA ligase I.
- Two replication forks are established at each origin and they move in opposite directions, so each origin creates a bidirectional replication unit or replicon.
- Fork progression is coordinated by the replisome which contains helicase, polymerases, sliding clamp and clamp loader, and accessory factors are associated.
- Ahead of the fork supercoiling is generated and this torsional strain is relieved by topoisomerases, which cleave and reseal DNA to allow fork movement.
- If obstacles (DNA lesions, tightly bound proteins) are encountered forks can stall, restart pathways are then engaged such as template switching or translesion synthesis, they act.
- The replisome components are dynamically exchanged sometimes, and replication proteins are recycled, making the process adaptable to local chromatin.
- As adjacent replicons expand, forks from neighboring origins eventually converge and termination occurs when two forks meet, processing of the junction is required.
- Termination junctions are processed and any remaining RNA primers or flaps are removed, this processing is done by nucleases and ligases to restore continuity.
- Daughter duplexes can become catenated as replication finishes, and type II topoisomerases (topo II) are used to decatenate by passing one duplex through another then resealing.
- The end-replication problem for linear chromosomes is encountered because terminal RNA primers cannot be fully replaced, leading to gradual shortening unless countered.
- Telomeres (repetitive sequences) are maintained by telomerase in germline/stem cells, telomerase extends 3’ ends using an RNA template, preventing loss of coding DNA.
- Nucleosomes are disassembled ahead of the fork and histones are recycled and newly synthesized histones are deposited by chaperones (CAF-1 etc.), chromatin is reassembled behind the fork.
- Replication timing is programed so that origins fire at different times (early vs late replicating domains), and timing is regulated by chromatin context and cell-type specific factors.
- Proofreading by polymerases and post-replicative mismatch repair (MMR) are used to correct errors, yet some mismatches escape and repair pathways act later.
- After replication, sister chromatids are held together by cohesion complexes until mitosis, cohesion establishment is coupled to replication and they are removed later for segregation.
- Checkpoints (ATR/Chk1 etc.) monitor fork integrity and DNA damage, and cell-cycle progression is paused if replication problems are detected, repair is then attempted.
- The overall rates vary: fork speeds are ~20–100 nt/sec in eukaryotes (varies by species and condition), and origin spacing is roughly 50–300 kb apart depending on cell type.
- Such as, in Saccharomyces cerevisiae origins are well-defined ARS sequences, while in metazoans origins are flexible and not always sequence-specific.
- Walking through steps: origins licensed → origins fired → helicase activation → bubble formation → bidirectional fork progression → leading/lagging synthesis → Okazaki processing → fork convergence → termination → decatenation → chromatin reassembly.
- Errors in initiation or termination are sometimes produced, causing re-replication or under-replication, and cells rely on checkpoints and repair to resolve these issues.
- The process is mostly highly regulated and accurate, but it’s messy in practice, it’s coordinated with chromatin, cell-cycle and repair systems, and they keep genome integrity.
Key Features of Bidirectional Replication
- Bidirectional Forks: Two replication forks form at the origin and move in opposite directions, enabling efficient replication of the entire genome.
- Leading and Lagging Strands: Continuous synthesis occurs on the leading strand, while discontinuous synthesis involving Okazaki fragments occurs on the lagging strand.
- Role of Topoisomerase: Prevents overwinding ahead of the replication fork and resolves daughter DNA molecules after replication.
4. Replication of the 5′ End of Linear Chromosomes (Telomere Replication)
Replication of the 5′ end of linear chromosomes is considered problematic, because RNA primers cannot be replaced at the extreme terminus, and progressive shortening is produced.
The end-replication problem is described as loss of terminal sequence when lagging strand synthesis leaves a gap after primer removal, and that gap are left unfilled.
Telomerase (TERT = telomerase reverse transcriptase; TERC = telomerase RNA component) extends the 3′ end by adding repeats, and telomerase extends the 3’ overhang actively using its RNA as template.
After extension by telomerase, the complementary 5′ end is synthesized by priming and conventional polymerases (primase/Pol α then Pol δ/Pol ε), and nicks are ligated to restore continuity.
A single-stranded 3′ overhang is maintained, and that overhang is used for repeated extension or for formation of a protective loop structure (t-loop), which is stabilized by shelterin proteins.
The protective function is provided by telomere-binding proteins (e.g., TRF1, TRF2) which are bound and regulate access of telomerase and repair factors, yet sometimes they block repair, confusing the machinery.
In many somatic cells telomerase activity is low or absent, so telomeres are progressively shortened with each division, leading to replicative senescence or genomic instability.
Alternative Lengthening of Telomeres (ALT) pathways are used by some cells/ tumours, where recombination-based mechanisms are employed to maintain telomere length, and they rely on HR proteins.
The sequence repeats are species-specific (Such as, TTAGGG repeats in vertebrates), and average lengths vary (e.g., 5–15 kb in humans at birth, then shortened with age), which is reported in ranges.
Telomere replication is coordinated with replication forks, and origin timing/ firing influence how telomeres are processed, yet forks near ends are often fragile and can stall.
Damage or incomplete replication at ends are sensed by DNA damage responses and checkpoint kinases, and repair or salvage pathways are activated to prevent end-to-end fusions.
In yeasts like Saccharomyces cerevisiae telomerase action and end processing were characterized experimentally, and proteins differ somewhat from vertebrates but function is conserved.
The maintenance is tightly regulated, telomerase is recruited by accessory factors, and its activity is modulated by cell type, cell-cycle stage, and epigenetic context.
Consequences of failure are seen as shortening → senescence → apoptosis or chromosomal instability (fusions, rearrangements), which impacts aging and cancer risk.
Walking along the short overview: primer problem → telomerase extension of 3′ → fill-in of complementary strand → protection by shelterin/t-loop → outcome (stable or shortened telomere).
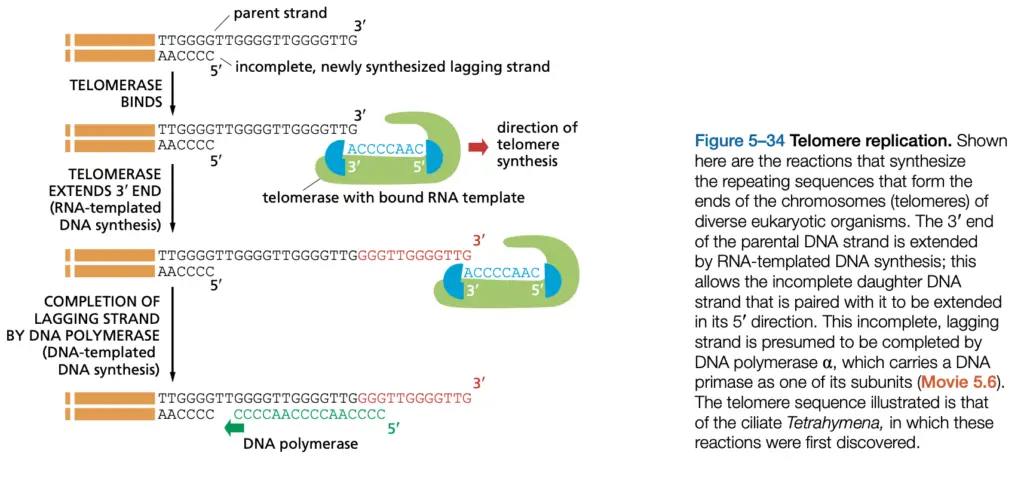
Steps of Telomere Replication
- The end-replication problem is described as a gap at the 5′ terminus after removal of RNA primers, and progressive shortening is caused.
- The replication fork is progressed toward the end, and RNA primer that initiated the final Okazaki fragment are removed, leaving a terminal gap.
- This 5′ gap are recognized as an unfillable gap by normal DNA polymerases, and conventional fill-in is prevented.
- Telomerase (a ribonucleoprotein, TERT = telomerase reverse transcriptase; TERC = telomerase RNA component) is recruited to the 3′ overhang, and binding is mediated by accessory factors.
- The 3′ overhang is extended by telomerase, which uses its internal RNA as template, and new repeats (Such as, TTAGGG) are added one repeat at a time.
- Telomerase activity is sometimes regulated by shelterin proteins ; they are bound to telomeres and modulate access, and sometimes they block telomerase.
- After several repeats are added, the extended 3′ end is used as a substrate for conventional priming, and primase (Pol α-primase) is recruited to make a short RNA primer.
- The short RNA primer provides a 3’-OH, and DNA polymerase (Pol δ / Pol ε depending on system) is used to extend the complementary strand toward the terminus.
- RNA primers are removed by RNase H and flap endonucleases (FEN1), and the resulting gaps are filled in by Pol δ, then nicks are sealed by DNA ligase I.
- A short 3′ single-stranded overhang is intentionally left, and that overhang is important for forming protective structures (t-loop) and for protein binding.
- The 3′ overhang invades the duplex telomeric repeats to form a T-loop, which is stabilized by shelterin complex components (TRF1, TRF2, POT1 etc.).
- Shelterin proteins are bound and end protection is provided, preventing the end from being mistaken as a DNA break by repair systems.
- In germline and stem cells telomerase is active and telomeres are elongated, while in most somatic cells telomerase is low/absent, so telomeres shorten with divisions.
- Alternative Lengthening of Telomeres (ALT) pathways are used by some cells/tumors, where recombination based mechanisms are employed to extend telomeres, and HR proteins are co-opted.
- The sequence repeat length is species-specific (Such as, TTAGGG in vertebrates), and lengths vary widely — e.g., 5–15 kb in humans at birth, then shortened with age.
- Telomerase is recruited in S/G2 phase often, and its action is coordinated with replication fork progression so that extension follows replication of nearby sequences.
- After telomerase detaches, the complementary strand synthesis is completed, and the telomere is processed to generate the proper overhang size.
- Errors in telomerase action or in primer removal are produced sometimes, causing deletions or aberrant ends, which may be repaired or lead to instability.
- The final protected end is maintained by protein complexes, and they are required to prevent end-to-end fusions and checkpoint activation.
- In yeast (e.g., Saccharomyces cerevisiae) proteins differ but the principle is conserved: telomerase extension → fill-in → protection.
- Fragmented summary: primer removed → 3′ overhang exposed → telomerase binds/extends → priming of complementary strand → fill-in by Pol δ → ligation → t-loop formation → shelterin protection.
- The process is mostly passive in its recognition steps, but telomerase sometimes acts actively to add repeats, and they together maintain chromosome integrity though not perfectly.
- https://www.onlinebiologynotes.com/mechanism-of-plasmid-replication-theta-and-rolling-circle-dna-replication/
- https://biology.stackexchange.com/questions/112036/telomerase-and-end-replication-in-eukaryote
- https://oertx.highered.texas.gov/courseware/lesson/1679/student-old/?task=2
- https://viralzone.expasy.org/1939
- Text Highlighting: Select any text in the post content to highlight it
- Text Annotation: Select text and add comments with annotations
- Comment Management: Edit or delete your own comments
- Highlight Management: Remove your own highlights
How to use: Simply select any text in the post content above, and you'll see annotation options. Login here or create an account to get started.